bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2590_orf2
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
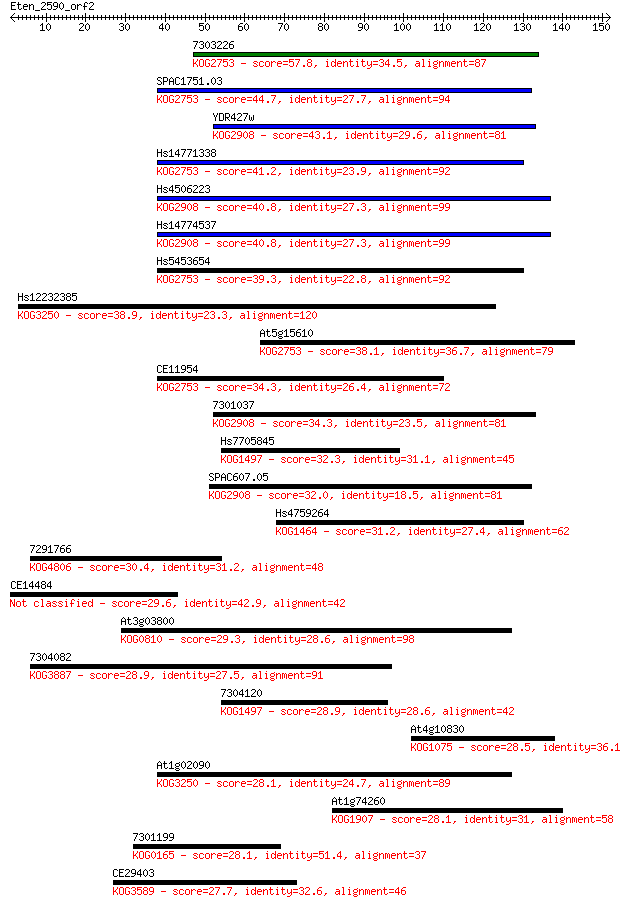
7303226 57.8 7e-09
SPAC1751.03 44.7 8e-05
YDR427w 43.1 2e-04
Hs14771338 41.2 8e-04
Hs4506223 40.8 9e-04
Hs14774537 40.8 9e-04
Hs5453654 39.3 0.003
Hs12232385 38.9 0.003
At5g15610 38.1 0.007
CE11954 34.3 0.090
7301037 34.3 0.091
Hs7705845 32.3 0.35
SPAC607.05 32.0 0.44
Hs4759264 31.2 0.77
7291766 30.4 1.5
CE14484 29.6 2.2
At3g03800 29.3 3.3
7304082 28.9 3.6
7304120 28.9 3.7
At4g10830 28.5 5.2
At1g02090 28.1 6.3
At1g74260 28.1 6.4
7301199 28.1 7.4
CE29403 27.7 7.8
> 7303226
Length=387
Score = 57.8 bits (138), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 53/99 (53%), Gaps = 12/99 (12%)
Query 47 TLQRRRLFSFKQLSDFS------------QLSEATAEQLVVQAIGQGLVEAKVDQINKLV 94
+++ RL +F QL++ S Q++E E V++ + LV A++DQ N+ V
Sbjct 274 NMKKMRLLTFMQLAESSPEMTFETLTKELQINEDEVEPFVIEVLKTKLVRARLDQANQKV 333
Query 95 HVRTAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESV 133
H+ + + R FG QWE++ L+ W + T+R+ SV
Sbjct 334 HISSTMHRTFGAPQWEQLRDLLQAWKENLSTVREGLTSV 372
> SPAC1751.03
Length=378
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 49/96 (51%), Gaps = 2/96 (2%)
Query 38 KLQLLWVAS--TLQRRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVH 95
K++LL +AS T S+ ++ ++ E E ++ I GLVE ++ Q+ K +
Sbjct 282 KMKLLTIASLATQAPNNTLSYGDVAKSLKIDENEVELWIIDVIRAGLVEGRMSQLTKTLS 341
Query 96 VRTAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKE 131
+ + R FG+ +W + +L W S++R + Q E
Sbjct 342 IHRSSYRVFGKHEWVALHEKLAKWGSSLRYMLQVME 377
> YDR427w
Length=393
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 42/81 (51%), Gaps = 0/81 (0%)
Query 52 RLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEE 111
R+ SF+ +S + L + E LV++AI GL++ +DQ+N+LV + R Q +
Sbjct 306 RMLSFEDISKATHLPKDNVEHLVMRAISLGLLKGSIDQVNELVTISWVQPRIISGDQITK 365
Query 112 VLGQLEHWASAVRTLRQAKES 132
+ +L W V L + E+
Sbjct 366 MKDRLVEWNDQVEKLGKKMEA 386
> Hs14771338
Length=374
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 41/92 (44%), Gaps = 0/92 (0%)
Query 38 KLQLLWVASTLQRRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVR 97
K++LL + SF + Q+ E V+ A+ +V K+DQ + V V
Sbjct 276 KMRLLTFMGMAVENKEISFDTMQQELQIGADDVEAFVIDAVRTKMVYCKIDQTQRKVVVS 335
Query 98 TAIQREFGRAQWEEVLGQLEHWASAVRTLRQA 129
+ R FG+ QW+++ L W + ++ +
Sbjct 336 HSTHRTFGKQQWQQLYDTLNAWKQNLNKVKNS 367
> Hs4506223
Length=376
Score = 40.8 bits (94), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 54/103 (52%), Gaps = 4/103 (3%)
Query 38 KLQLLWVAS-TLQR---RRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKL 93
K+QLL + T R R +F++++ ++++ E LV++A+ GLV+ +D+++K
Sbjct 271 KIQLLCLMEMTFTRPANHRQLTFEEIAKSAKITVNEVELLVMKALSVGLVKGSIDEVDKR 330
Query 94 VHVRTAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESVKHS 136
VH+ R Q + + +LE W + V+++ E H
Sbjct 331 VHMTWVQPRVLDLQQIKGMKDRLEFWCTDVKSMEMLVEHQAHD 373
> Hs14774537
Length=376
Score = 40.8 bits (94), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 54/103 (52%), Gaps = 4/103 (3%)
Query 38 KLQLLWVAS-TLQR---RRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKL 93
K+QLL + T R R +F++++ ++++ E LV++A+ GLV+ +D+++K
Sbjct 271 KIQLLCLMEMTFTRPANHRQLTFEEIAKSAKITVNEVELLVMKALSVGLVKGSIDEVDKR 330
Query 94 VHVRTAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESVKHS 136
VH+ R Q + + +LE W + V+++ E H
Sbjct 331 VHMTWVQPRVLDLQQIKGMKDRLEFWCTDVKSMEMLVEHQAHD 373
> Hs5453654
Length=374
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/92 (22%), Positives = 41/92 (44%), Gaps = 0/92 (0%)
Query 38 KLQLLWVASTLQRRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVR 97
K++LL + SF + Q+ E V+ A+ +V K+DQ + V V
Sbjct 276 KMRLLTFMGMAIENKEISFDTMQQELQIGADDVEAFVIDAVRTKMVYCKIDQTQRKVVVS 335
Query 98 TAIQREFGRAQWEEVLGQLEHWASAVRTLRQA 129
+ R FG+ +W+++ L W + ++ +
Sbjct 336 HSTHRTFGKQRWQQLYDTLNAWKQNLNKVKNS 367
> Hs12232385
Length=264
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/123 (22%), Positives = 52/123 (42%), Gaps = 12/123 (9%)
Query 3 LNILFFSIFCKSSANRALSSSKSSSPSTPKCST--NTKLQLLWVASTLQRRRLFSFK-QL 59
LN+ + + AN+ S P+ ST KL+ L + S R + + L
Sbjct 67 LNLFAYGTYPDYIANKE---------SLPELSTAQQNKLKHLTIVSLASRMKCIPYSVLL 117
Query 60 SDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHW 119
D + E L+++A+ +++ K+DQ N+L+ V I R+ + ++ L W
Sbjct 118 KDLEMRNLRELEDLIIEAVYTDIIQGKLDQRNQLLEVDFCIGRDIRKKDINNIVKTLHEW 177
Query 120 ASA 122
Sbjct 178 CDG 180
> At5g15610
Length=442
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 46/108 (42%), Gaps = 29/108 (26%)
Query 64 QLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRT------------------------- 98
Q+++ E VV+AI LV K+DQ+N++V VR
Sbjct 325 QVNDEEVELWVVKAITAKLVACKMDQMNQVVIVRQVSNLLLFRFICVNPKSHILVLHICS 384
Query 99 ----AIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESVKHSAQPFET 142
+REFG+ QW+ + +L W VR + ES K + + +T
Sbjct 385 CISRCAEREFGQKQWQSLRTKLAAWRDNVRNVISTIESNKATEEGTQT 432
> CE11954
Length=390
Score = 34.3 bits (77), Expect = 0.090, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 38/73 (52%), Gaps = 1/73 (1%)
Query 38 KLQLLWVASTLQRRRLFSFKQLSD-FSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHV 96
K++LL + S + + S +L+ L++ T E+ V+ AI + K++++ + + V
Sbjct 287 KIRLLTLMSLAEEKNEISLDELAKQLDILADETLEEFVIDAIQVNAISGKINEMARTLIV 346
Query 97 RTAIQREFGRAQW 109
+ R FG QW
Sbjct 347 SSYQHRRFGTEQW 359
> 7301037
Length=382
Score = 34.3 bits (77), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 19/81 (23%), Positives = 40/81 (49%), Gaps = 0/81 (0%)
Query 52 RLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEE 111
R SF ++ ++L E L+++A+ LV ++DQ+ +V++ R R+Q
Sbjct 294 RAISFTDIAQETKLPAKEVELLIMKALALDLVRGEIDQVAGVVNMSWVQPRVLNRSQIVG 353
Query 112 VLGQLEHWASAVRTLRQAKES 132
+ L+ W A+ + + E+
Sbjct 354 MASTLDTWMGAITNMEKLMEN 374
> Hs7705845
Length=405
Score = 32.3 bits (72), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 54 FSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRT 98
+F++L ++ A AE++ Q I +G + +DQI+ +VH T
Sbjct 318 ITFEELGALLEIPAAKAEKIASQMITEGRMNGFIDQIDGIVHFET 362
> SPAC607.05
Length=381
Score = 32.0 bits (71), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 15/81 (18%), Positives = 39/81 (48%), Gaps = 0/81 (0%)
Query 51 RRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWE 110
+R +F ++ +++ E L+++A+ GL+ +D++ ++V + + R +Q
Sbjct 293 QRTLTFDTIARATRIPSNEVELLIMRALSVGLITGVIDEVTQIVTISSVQSRILNHSQIA 352
Query 111 EVLGQLEHWASAVRTLRQAKE 131
+ +L W ++ L E
Sbjct 353 SMESRLREWNQNIKNLSNVVE 373
> Hs4759264
Length=443
Score = 31.2 bits (69), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 33/62 (53%), Gaps = 7/62 (11%)
Query 68 ATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASAVRTLR 127
A E L+VQ I + ++DQ+N+L+ + ++ G A++ L+ W + + +L
Sbjct 383 ADVESLLVQCILDNTIHGRIDQVNQLLELD---HQKRGGARY----TALDKWTNQLNSLN 435
Query 128 QA 129
QA
Sbjct 436 QA 437
> 7291766
Length=462
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 24/48 (50%), Gaps = 5/48 (10%)
Query 6 LFFSIFCKSSANRALSSSKSSSPSTPKCSTNTKLQLLWVASTLQRRRL 53
+ F+ FC+ A+S+ P TP C+ L+ +W A + RRL
Sbjct 113 MIFTNFCQ-----AVSAYTDQQPMTPSCAAGGPLEGIWGAPPQRERRL 155
> CE14484
Length=404
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 26/44 (59%), Gaps = 2/44 (4%)
Query 1 IKLNILFFSIFCKSSANRALSSSKSSSPSTPKC--STNTKLQLL 42
I++NI+ +I C S+N+ LS S + KC NTKL+LL
Sbjct 46 IQMNIIMITISCIHSSNQLLSDFAFRSQAMLKCHPGFNTKLELL 89
> At3g03800
Length=306
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 47/111 (42%), Gaps = 17/111 (15%)
Query 29 STPKCSTNTKLQLLWVASTLQRRRLFSFKQLSDFSQLSE-------ATAEQLVVQAIGQG 81
+ P C T + A+T+ ++ F K +S+F L + E+ V GQ
Sbjct 112 TKPGCGKGTGVDRTRTATTIAVKKKFKDK-ISEFQTLRQNIQQEYREVVERRVFTVTGQ- 169
Query 82 LVEAKVDQINKLVHVRTAIQ------REFGRAQWEEVLGQLEHWASAVRTL 126
A + I++L+ + Q RE GR Q + L +++ AVR L
Sbjct 170 --RADEEAIDRLIETGDSEQIFQKAIREQGRGQIMDTLAEIQERHDAVRDL 218
> 7304082
Length=387
Score = 28.9 bits (63), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 25/99 (25%), Positives = 41/99 (41%), Gaps = 8/99 (8%)
Query 6 LFFSIFCKSSANRALSSSKSSSPSTPKCSTNTKLQLL--------WVASTLQRRRLFSFK 57
L F I K N AL+ K++ K + K ++ W L +R F
Sbjct 119 LVFVIDAKDDYNEALTKFKNTVLQAYKVNKRIKFEVFIHKVISMSWALIVLITQRHMLFS 178
Query 58 QLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHV 96
Q+ S S+ +++ + Q L EA +DQI+ H+
Sbjct 179 QVDGISDDSKMESQRDIHQRSSDDLNEAGLDQIHLSFHL 217
> 7304120
Length=407
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 12/42 (28%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 54 FSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVH 95
+F++L + AE++ Q I +G + +DQI+ +VH
Sbjct 326 ITFEELGALLDIPAVKAEKIASQMITEGRMNGHIDQISAIVH 367
> At4g10830
Length=1294
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Query 102 REFGRAQWEEVL-GQLEHWASAVRTLRQAKESVKHSA 137
+ + +A W + + GQ +H VRT RQA +KH +
Sbjct 614 KTYVKAGWNKAINGQRKHLPDQVRTCRQAMAKLKHKS 650
> At1g02090
Length=260
Score = 28.1 bits (61), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 41/92 (44%), Gaps = 4/92 (4%)
Query 38 KLQLLWVASTLQRRRLFSFKQLS---DFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLV 94
KL+ L V + + ++ + L D S + E + L+ + + G+V K+DQ+ +
Sbjct 94 KLKQLTVLTLAESNKVLPYDTLMVELDVSNVRE-LEDFLINECMYAGIVRGKLDQLKRCF 152
Query 95 HVRTAIQREFGRAQWEEVLGQLEHWASAVRTL 126
V A R+ Q +L L +W + L
Sbjct 153 EVPFAAGRDLRPGQLGNMLHTLSNWLNTSENL 184
> At1g74260
Length=1387
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 24/58 (41%), Gaps = 5/58 (8%)
Query 82 LVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESVKHSAQP 139
L+E KVD I L + F R WE+ QLE + KE +K +P
Sbjct 1046 LIEVKVDGITHLSE-----KTSFLRDMWEDTSFQLEKLQRLASCVEMEKEGLKFRHEP 1098
> 7301199
Length=1954
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 23/40 (57%), Gaps = 3/40 (7%)
Query 32 KCSTNTKLQLLWVASTLQRR---RLFSFKQLSDFSQLSEA 68
K N+ LQL A TLQRR RL KQL ++QL +A
Sbjct 1404 KRERNSFLQLRQAAITLQRRYRARLNMIKQLKSYAQLKQA 1443
> CE29403
Length=819
Score = 27.7 bits (60), Expect = 7.8, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 29/46 (63%), Gaps = 1/46 (2%)
Query 27 SPSTPKCSTNTKLQLLWVASTLQRRRLFSFKQLSDFSQLSEATAEQ 72
+P +P CS+ + Q ++ + RR+ S QLS+ S+L E+T+++
Sbjct 199 APMSPVCSSTPRSQRMYRKNPKYRRQFGSSLQLSE-SRLEESTSQE 243
Lambda K H
0.317 0.126 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1888713112
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40