bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2573_orf1
Length=180
Score E
Sequences producing significant alignments: (Bits) Value
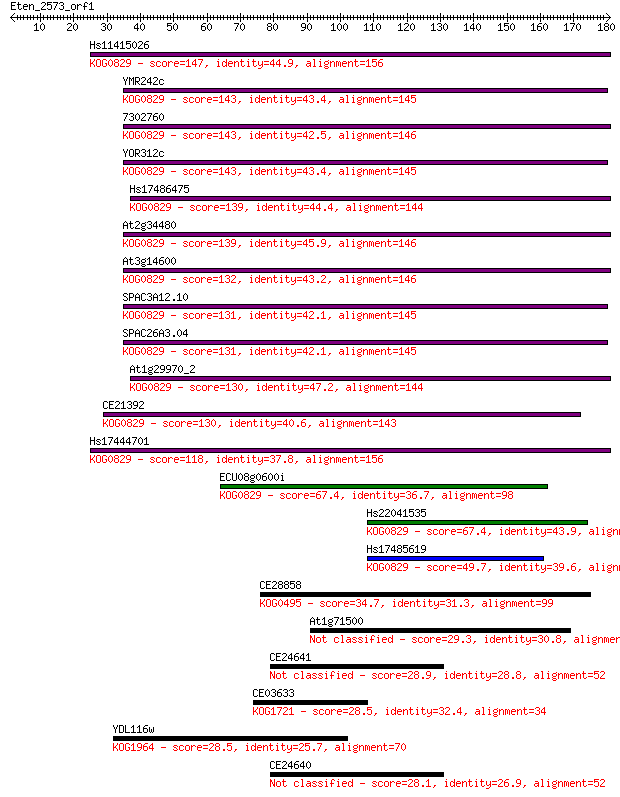
Hs11415026 147 1e-35
YMR242c 143 2e-34
7302760 143 2e-34
YOR312c 143 2e-34
Hs17486475 139 3e-33
At2g34480 139 3e-33
At3g14600 132 3e-31
SPAC3A12.10 131 8e-31
SPAC26A3.04 131 8e-31
At1g29970_2 130 1e-30
CE21392 130 2e-30
Hs17444701 118 8e-27
ECU08g0600i 67.4 1e-11
Hs22041535 67.4 1e-11
Hs17485619 49.7 3e-06
CE28858 34.7 0.11
At1g71500 29.3 4.4
CE24641 28.9 5.2
CE03633 28.5 6.6
YDL116w 28.5 7.8
CE24640 28.1 9.8
> Hs11415026
Length=176
Score = 147 bits (370), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 70/156 (44%), Positives = 100/156 (64%), Gaps = 8/156 (5%)
Query 25 MKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLR 84
MKA TL R+++VVGR P P YR+R+FA N V+A SRFWY + QL+
Sbjct 1 MKASGTL----REYKVVGRCLPTPKCHTP----PLYRMRIFAPNHVVAKSRFWYFVSQLK 52
Query 85 KIKRAHGEVLEVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAE 144
K+K++ GE++ ++ K KNFG+WLRYDSR+G+HNMY+E RD++ A Q +
Sbjct 53 KMKKSSGEIVYCGQVFEKSPLRVKNFGIWLRYDSRSGTHNMYREYRDLTTAGAVTQCYRD 112
Query 145 MAGRHRALASNIQIIRIVQLKGSECRRPHVQQMLSS 180
M RHRA A +IQI+++ ++ S+CRRP V+Q S
Sbjct 113 MGARHRARAHSIQIMKVEEIAASKCRRPAVKQFHDS 148
> YMR242c
Length=180
Score = 143 bits (360), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 63/146 (43%), Positives = 102/146 (69%), Gaps = 5/146 (3%)
Query 35 IRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVL 94
+++QV+GR+ P + +R+R+FA+NEV+A SR+WY L++L K+K+A GE++
Sbjct 12 FKEYQVIGRRLPTESVPEPKL----FRMRIFASNEVIAKSRYWYFLQKLHKVKKASGEIV 67
Query 95 EVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMAGRHRALAS 154
+ +IN T KNFGVW+RYDSR+G+HNMYKE+RD+S AA + +MA RHRA
Sbjct 68 SINQINEAHPTKVKNFGVWVRYDSRSGTHNMYKEIRDVSRVAAVETLYQDMAARHRARFR 127
Query 155 NIQIIRIVQL-KGSECRRPHVQQMLS 179
+I I+++ ++ K ++ +R +V+Q L+
Sbjct 128 SIHILKVAEIEKTADVKRQYVKQFLT 153
> 7302760
Length=177
Score = 143 bits (360), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 62/146 (42%), Positives = 96/146 (65%), Gaps = 4/146 (2%)
Query 35 IRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVL 94
+++++VVGR+ P+ TP Y++R+FA + ++A SRFWY LRQL+K K+ GE++
Sbjct 7 LKEYEVVGRKLPSEKEPQ----TPLYKMRIFAPDNIVAKSRFWYFLRQLKKFKKTTGEIV 62
Query 95 EVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMAGRHRALAS 154
+ ++ KNFG+WLRYDSR+G+HNMY+E RD++ A Q +M RHRA A
Sbjct 63 SIKQVYETSPVKIKNFGIWLRYDSRSGTHNMYREYRDLTVGGAVTQCYRDMGARHRARAH 122
Query 155 NIQIIRIVQLKGSECRRPHVQQMLSS 180
+IQII++ + ++ RR HV+Q S
Sbjct 123 SIQIIKVDSIPAAKTRRVHVKQFHDS 148
> YOR312c
Length=174
Score = 143 bits (360), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 63/146 (43%), Positives = 102/146 (69%), Gaps = 5/146 (3%)
Query 35 IRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVL 94
+++QV+GR+ P + +R+R+FA+NEV+A SR+WY L++L K+K+A GE++
Sbjct 6 FKEYQVIGRRLPTESVPEPKL----FRMRIFASNEVIAKSRYWYFLQKLHKVKKASGEIV 61
Query 95 EVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMAGRHRALAS 154
+ +IN T KNFGVW+RYDSR+G+HNMYKE+RD+S AA + +MA RHRA
Sbjct 62 SINQINEAHPTKVKNFGVWVRYDSRSGTHNMYKEIRDVSRVAAVETLYQDMAARHRARFR 121
Query 155 NIQIIRIVQL-KGSECRRPHVQQMLS 179
+I I+++ ++ K ++ +R +V+Q L+
Sbjct 122 SIHILKVAEIEKTADVKRQYVKQFLT 147
> Hs17486475
Length=176
Score = 139 bits (350), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 64/144 (44%), Positives = 92/144 (63%), Gaps = 4/144 (2%)
Query 37 QFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVLEV 96
++QVVGR PA P YR+R+FA N V+A S FWY + QL+K+K++ GE++
Sbjct 9 EYQVVGRCLPAPKCHTP----PLYRMRIFAPNHVVAKSHFWYFVSQLKKLKKSSGEIVYC 64
Query 97 VEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMAGRHRALASNI 156
++ KR KNFG+WLRYDSR G+HN+Y+E RD++ A + +M RHRA A +I
Sbjct 65 GQVFEKRPLRVKNFGIWLRYDSRRGTHNIYREYRDLTTAGAVTKCYRDMGARHRARAHSI 124
Query 157 QIIRIVQLKGSECRRPHVQQMLSS 180
QI ++ + S+CRRP V+Q S
Sbjct 125 QIRKVEDIAASKCRRPTVKQFHDS 148
> At2g34480
Length=178
Score = 139 bits (350), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 67/146 (45%), Positives = 89/146 (60%), Gaps = 4/146 (2%)
Query 35 IRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVL 94
Q+QVVGR P YR++L+A NEV A S+FWY LR+L+K+K+++G++L
Sbjct 6 FHQYQVVGRALPTEKDVQPKI----YRMKLWATNEVRAKSKFWYFLRKLKKVKKSNGQML 61
Query 95 EVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMAGRHRALAS 154
+ EI K T KNFG+WLRY SRTG HNMYKE RD + N A Q+ EMA RHR
Sbjct 62 AINEIYEKNPTTIKNFGIWLRYQSRTGYHNMYKEYRDTTLNGAVEQMYTEMASRHRVRFP 121
Query 155 NIQIIRIVQLKGSECRRPHVQQMLSS 180
IQII+ + C+R +Q +S
Sbjct 122 CIQIIKTATVPAKLCKRESTKQFHNS 147
> At3g14600
Length=178
Score = 132 bits (332), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 63/146 (43%), Positives = 87/146 (59%), Gaps = 4/146 (2%)
Query 35 IRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVL 94
Q+QVVGR P YR++L+ NEV A S+FWY +R+L+K+K+++G++L
Sbjct 6 FHQYQVVGRALPTENDEHPKI----YRMKLWGRNEVCAKSKFWYFMRKLKKVKKSNGQML 61
Query 95 EVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMAGRHRALAS 154
+ EI K T KN+G+WLRY SRTG HNMYKE RD + N Q+ EMA RHR
Sbjct 62 AINEIFEKNPTTIKNYGIWLRYQSRTGYHNMYKEYRDTTLNGGVEQMYTEMASRHRVRFP 121
Query 155 NIQIIRIVQLKGSECRRPHVQQMLSS 180
IQII+ + C+R +Q +S
Sbjct 122 CIQIIKTATVPAKLCKREITKQFHNS 147
> SPAC3A12.10
Length=176
Score = 131 bits (329), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 61/146 (41%), Positives = 98/146 (67%), Gaps = 5/146 (3%)
Query 35 IRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVL 94
++++QVVGR+ P +R+RLFA NE +A SR+WY L+ + K+K+A GE++
Sbjct 3 LKEYQVVGRKVPTEHEPVPKL----FRMRLFAPNESVAKSRYWYFLKMINKVKKATGEIV 58
Query 95 EVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMAGRHRALAS 154
+ EI+ + K FG+W+RYDSR+G+HNMYKE RD + A + A+MA RHRA
Sbjct 59 AINEISEPKPLKAKVFGIWIRYDSRSGTHNMYKEFRDTTRVGAVEAMYADMAARHRARFR 118
Query 155 NIQIIRIVQL-KGSECRRPHVQQMLS 179
+I+I+++V++ K + RR +V+Q+L+
Sbjct 119 SIRILKVVEVEKKEDVRRNYVKQLLN 144
> SPAC26A3.04
Length=176
Score = 131 bits (329), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 61/146 (41%), Positives = 98/146 (67%), Gaps = 5/146 (3%)
Query 35 IRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVL 94
++++QVVGR+ P +R+RLFA NE +A SR+WY L+ + K+K+A GE++
Sbjct 3 LKEYQVVGRKVPTEHEPVPKL----FRMRLFAPNESVAKSRYWYFLKMINKVKKATGEIV 58
Query 95 EVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMAGRHRALAS 154
+ EI+ + K FG+W+RYDSR+G+HNMYKE RD + A + A+MA RHRA
Sbjct 59 AINEISEPKPLKAKVFGIWIRYDSRSGTHNMYKEFRDTTRVGAVEAMYADMAARHRARFR 118
Query 155 NIQIIRIVQL-KGSECRRPHVQQMLS 179
+I+I+++V++ K + RR +V+Q+L+
Sbjct 119 SIRILKVVEVEKKEDVRRNYVKQLLN 144
> At1g29970_2
Length=184
Score = 130 bits (328), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 68/156 (43%), Positives = 90/156 (57%), Gaps = 16/156 (10%)
Query 37 QFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHGEVLEV 96
Q+QVVGR P YR++L+A NEVLA S+FWY LR+ +K+K+++G++L +
Sbjct 2 QYQVVGRALPTEKDEQPKI----YRMKLWATNEVLAKSKFWYYLRRQKKVKKSNGQMLAI 57
Query 97 VE------------INPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAE 144
E I K T KNFG+WLRY SRTG HNMYKE RD + N A Q+ E
Sbjct 58 NEVLAVGSEFDLFSIFEKNPTTIKNFGIWLRYQSRTGYHNMYKEYRDTTLNGAVEQMYTE 117
Query 145 MAGRHRALASNIQIIRIVQLKGSECRRPHVQQMLSS 180
MA RHR IQII+ + S C+R +Q +S
Sbjct 118 MASRHRVRFPCIQIIKTATVPASLCKRESTKQFHNS 153
> CE21392
Length=180
Score = 130 bits (326), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 58/143 (40%), Positives = 89/143 (62%), Gaps = 4/143 (2%)
Query 29 KTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKR 88
K L + + ++ VVGR+ P TP +++++FA N V+A SRFWY + LR++K+
Sbjct 4 KALGETLNEYVVVGRKIPTEKEPV----TPIWKMQIFATNHVIAKSRFWYFVSMLRRVKK 59
Query 89 AHGEVLEVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMAGR 148
A+GE+L + ++ K KN+GVWL+YDSRTG HNMY+E RD + A Q +M R
Sbjct 60 ANGEILSIKQVFEKNPGTVKNYGVWLKYDSRTGHHNMYREYRDTTVAGAVTQCYRDMGAR 119
Query 149 HRALASNIQIIRIVQLKGSECRR 171
HRA A I I+++ +K + +R
Sbjct 120 HRAQADRIHILKVQTVKAEDTKR 142
> Hs17444701
Length=175
Score = 118 bits (295), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 59/156 (37%), Positives = 90/156 (57%), Gaps = 8/156 (5%)
Query 25 MKADKTLQQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLR 84
MKA T Q+ ++VV P P Y +++FA N V+A FWY L QL+
Sbjct 1 MKALGTQQE----YKVVCHCLPTPKCHTL----PLYHMQIFAPNHVVAKFHFWYFLSQLK 52
Query 85 KIKRAHGEVLEVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAE 144
K+K++ GE + ++ K KNFG+WLRYDSR+ +HNMY+E RD++ A Q +
Sbjct 53 KMKKSSGETVNCGQVFEKYPLWVKNFGIWLRYDSRSSTHNMYREYRDLTTMGAVTQCYQD 112
Query 145 MAGRHRALASNIQIIRIVQLKGSECRRPHVQQMLSS 180
M ++RA A+ IQI+++ ++ S+C P V+Q S
Sbjct 113 MGTQYRARANFIQIMKVEEIAASKCWWPVVKQFHDS 148
> ECU08g0600i
Length=173
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/98 (36%), Positives = 54/98 (55%), Gaps = 0/98 (0%)
Query 64 LFAANEVLAVSRFWYLLRQLRKIKRAHGEVLEVVEINPKRATNPKNFGVWLRYDSRTGSH 123
+F N V A ++F+ +L + KIK G +++ EI +N+G+ Y SR+G H
Sbjct 31 IFTKNHVFAKAKFFKILEKKYKIKATKGLIIDCKEIPEPSFKEVQNYGIRFVYRSRSGVH 90
Query 124 NMYKEVRDISENAAAAQVLAEMAGRHRALASNIQIIRI 161
N YKE R IS+ A VL E+AGR + S + II +
Sbjct 91 NAYKECRAISKCWAIDHVLRELAGRQKLKRSAVDIISV 128
> Hs22041535
Length=111
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 108 KNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMAGRHRALASNIQIIRIVQLKGS 167
KNFG+WL YDS++G+HN+Y+E RD++ A Q +HRALA +IQI++ Q
Sbjct 26 KNFGIWLGYDSQSGTHNLYREYRDLTPVGAVTQCYRNTGAQHRALAQSIQIMKRFQDSKI 85
Query 168 ECRRPH 173
+ PH
Sbjct 86 QFPLPH 91
> Hs17485619
Length=230
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 21/53 (39%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 108 KNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMAGRHRALASNIQIIR 160
KNF +WL ++S +G+HNMY+E D + A Q EM + A IQI++
Sbjct 112 KNFSLWLHHNSCSGTHNMYQEYLDSATGGAVTQYYREMGAQQCGWAHWIQIVK 164
> CE28858
Length=968
Score = 34.7 bits (78), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 50/108 (46%), Gaps = 18/108 (16%)
Query 76 FWYLLRQLRKIKRAHGEVLEV-VEINPKRATNPKNFGVWL---RYDSRTGSHNMYKE--V 129
W LL +L + G++++ V++ R NPKN +WL R++ R G M KE
Sbjct 772 LWILLVRLEE---KAGQIVKARVDLEKARLRNPKNDDLWLESVRFEQRVGCPEMAKERMS 828
Query 130 RDISENAAAAQVLAE---MAGRHRALASNIQIIRIVQLKGSECRRPHV 174
R + E + ++ AE M G H A +I ++ + PHV
Sbjct 829 RALQECEGSGKLWAEAIWMEGPHGRRAKSIDALKKCE------HNPHV 870
> At1g71500
Length=287
Score = 29.3 bits (64), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 41/84 (48%), Gaps = 10/84 (11%)
Query 91 GEVLEVVEINPK-RATNP---KNFGVWLRYDSRTGSHNMYKEVRDISENAAAAQVLAEMA 146
GE+ E NP R P K F ++YD N+Y +RD + AAA+++
Sbjct 156 GEIREWYPKNPVLRVLTPALRKLFVYPVKYDE----ENIYISIRDSGKTEAAAEIVFSGK 211
Query 147 GRHRALASNIQI--IRIVQLKGSE 168
+ A+N+ + +R++ +GSE
Sbjct 212 AQPGLTATNVNVDEVRMIVDEGSE 235
> CE24641
Length=308
Score = 28.9 bits (63), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 79 LLRQLRKIKRAHGEVLEVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVR 130
L R+L K + G+ +E+ + T+ FG + + GSHN +K +R
Sbjct 6 LCRKLVKFETESGKKVELDAVYEDSLTSGSPFGTVVAFHGSPGSHNDFKYIR 57
> CE03633
Length=2251
Score = 28.5 bits (62), Expect = 6.6, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 74 SRFWYLLRQLRKIKRAHGEVLEVVEINPKRATNP 107
+RF Y +R+ +KI + ++EV++ +P A P
Sbjct 933 ARFVYQMREFQKINNMNDRLIEVLKTDPAAAATP 966
> YDL116w
Length=726
Score = 28.5 bits (62), Expect = 7.8, Method: Composition-based stats.
Identities = 18/70 (25%), Positives = 33/70 (47%), Gaps = 1/70 (1%)
Query 32 QQPIRQFQVVGRQRPAAAAAAAAAATPAYRLRLFAANEVLAVSRFWYLLRQLRKIKRAHG 91
Q PI F ++ R AA A A + ++ + +RFW+L+ L + A
Sbjct 30 QNPIDPFNIIREFRSAAGQLALDLANSGDESNVISSKDWELEARFWHLVELLLVFRNADL 89
Query 92 EVLEVVEINP 101
+ L+ +E++P
Sbjct 90 D-LDEMELHP 98
> CE24640
Length=308
Score = 28.1 bits (61), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 79 LLRQLRKIKRAHGEVLEVVEINPKRATNPKNFGVWLRYDSRTGSHNMYKEVR 130
LL++L K + + + +E+ + T+ FG + + GSHN +K +R
Sbjct 14 LLKKLVKFQAENEQFVELEAVYEDSITSGSPFGTVVAFHGSPGSHNDFKYIR 65
Lambda K H
0.323 0.132 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2806646388
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40