bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
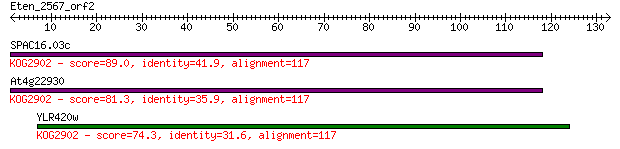
Query= Eten_2567_orf2
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
SPAC16.03c 89.0 2e-18
At4g22930 81.3 5e-16
YLR420w 74.3 5e-14
> SPAC16.03c
Length=337
Score = 89.0 bits (219), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 49/117 (41%), Positives = 66/117 (56%), Gaps = 9/117 (7%)
Query 1 LQVVREGHPQFFLGSDSAPHPRHKKDSYPPAAGVFTQPFLLAYLADTFARAGCLDKLQSF 60
++ +P+FF GSDSAPHPR K PPAAGVFTQPF +YLA+ F + G LD L+ F
Sbjct 225 IEAATSKNPKFFFGSDSAPHPRSSKLKTPPAAGVFTQPFAASYLAEVFDKEGRLDALKDF 284
Query 61 ACENAAAFFGLPKKELVEGEDCVVLEQTPCRIPNVVCGAEGTSTDVVPFLAGHELGY 117
AC F+ +P L E +VL++ R+P E + D+VPF L +
Sbjct 285 ACIFGRKFYCIP---LDFKESNIVLKKESFRVP------ESVANDLVPFHPNEVLQW 332
> At4g22930
Length=377
Score = 81.3 bits (199), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 42/117 (35%), Positives = 66/117 (56%), Gaps = 8/117 (6%)
Query 1 LQVVREGHPQFFLGSDSAPHPRHKKDSYPPAAGVFTQPFLLAYLADTFARAGCLDKLQSF 60
++ V G +FFLG+DSAPH R +K+S AG+++ P L+ A F AG LDKL++F
Sbjct 265 VKAVTSGSKKFFLGTDSAPHERSRKESSCGCAGIYSAPIALSLYAKVFDEAGALDKLEAF 324
Query 61 ACENAAAFFGLPKKELVEGEDCVVLEQTPCRIPNVVCGAEGTSTDVVPFLAGHELGY 117
N F+GLP+ + L+++P ++P+V G ++VP AG L +
Sbjct 325 TSFNGPDFYGLPR-----NSSKITLKKSPWKVPDVFNFPFG---EIVPMFAGETLQW 373
> YLR420w
Length=364
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 37/118 (31%), Positives = 64/118 (54%), Gaps = 4/118 (3%)
Query 7 GHPQFFLGSDSAPHPRHKKDSYPPA-AGVFTQPFLLAYLADTFARAGCLDKLQSFACENA 65
G P FF GSDSAPHP K +Y AGV++Q F + Y+A F L+ L+ F +
Sbjct 249 GKPYFFFGSDSAPHPVQNKANYEGVCAGVYSQSFAIPYIAQVFEEQNALENLKGFVSDFG 308
Query 66 AAFFGLPKKELVEGEDCVVLEQTPCRIPNVVCGAEGTSTDVVPFLAGHELGYTLKVVP 123
+F+ + E+ + ++ ++ IP V+ ++G ++PF AG +L ++++ P
Sbjct 309 ISFYEVKDSEVASSDKAILFKKEQV-IPQVI--SDGKDISIIPFKAGDKLSWSVRWEP 363
Lambda K H
0.322 0.138 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1319765976
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40