bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2551_orf4
Length=277
Score E
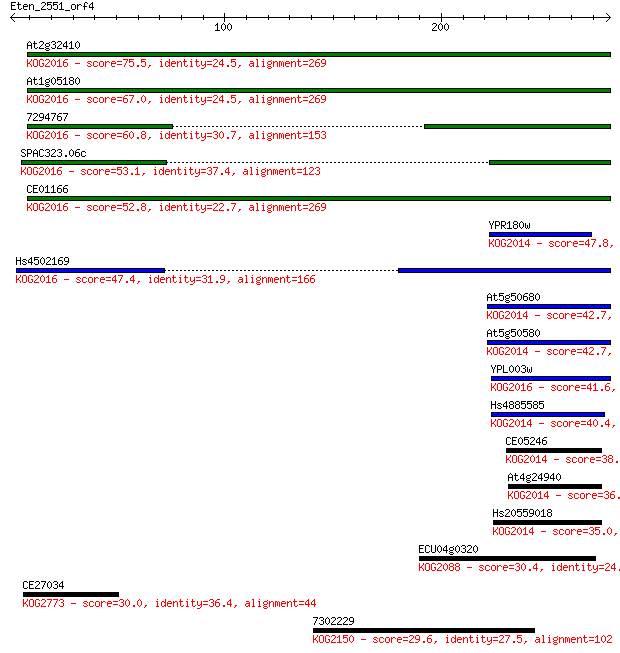
Sequences producing significant alignments: (Bits) Value
At2g32410 75.5 1e-13
At1g05180 67.0 4e-11
7294767 60.8 3e-09
SPAC323.06c 53.1 6e-07
CE01166 52.8 7e-07
YPR180w 47.8 3e-05
Hs4502169 47.4 3e-05
At5g50680 42.7 9e-04
At5g50580 42.7 9e-04
YPL003w 41.6 0.002
Hs4885585 40.4 0.004
CE05246 38.9 0.013
At4g24940 36.2 0.074
Hs20559018 35.0 0.17
ECU04g0320 30.4 3.8
CE27034 30.0 5.0
7302229 29.6 8.1
> At2g32410
Length=523
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 66/274 (24%), Positives = 113/274 (41%), Gaps = 74/274 (27%)
Query 9 DFAADTKSYLELQKIYNEKAERD----ERRLRELLGQLDVHPSAKLNVSQEEIKCFCKNA 64
D + T+ Y+ LQKIY+ KAE D E+R++ +L ++ PS+ +S+ IK FCKNA
Sbjct 319 DMISSTEHYINLQKIYHSKAEADFLSMEQRVKSILVKVGQDPSS---ISKPTIKSFCKNA 375
Query 65 VFFKVTQYPKITGAFLGSTSVELLAAAAREMQQAVPEELKLSPNADPDLFADPNFSVYEE 124
KV +Y I F ++ EL AD N
Sbjct 376 RKLKVCRYRTIEDEFKSPSTTEL-----------------------HKYLADEN------ 406
Query 125 NGFLHLPWYAAALACRAAAAKLGRFPGSHPKPSAAAATAATGEAAAAATGGAAAARSVEE 184
Y+ A+ + RF G++ K G+ + A+ +++
Sbjct 407 --------YSGAIGFYILLRAVDRFAGTYKK--------FPGQFDGSTDEDASQLKTIAL 450
Query 185 AIKQQQELVEKDSAAVLLELEEMEKAVGCRLFA-AAKVVQQLVSYRGSEFPATAALVGAV 243
++ L EM GC + ++ ++ + +E AAL+G +
Sbjct 451 SL-----------------LSEM----GCDGYELQEELYNEMCRFGAAEIHVVAALIGGI 489
Query 244 AAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
+QE IKL+ K+F P TF++NG+ + ++ L
Sbjct 490 TSQEVIKLITKQFVPKRGTFIFNGIDHKSQSLTL 523
> At1g05180
Length=540
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 66/274 (24%), Positives = 110/274 (40%), Gaps = 74/274 (27%)
Query 9 DFAADTKSYLELQKIYNEKAERD----ERRLRELLGQLDVHPSAKLNVSQEEIKCFCKNA 64
D + T+ Y+ LQKIY KAE D E R++ +L ++ PS+ + + IK FCKNA
Sbjct 336 DMTSSTEHYINLQKIYLAKAEADFLVIEERVKNILKKIGRDPSS---IPKPTIKSFCKNA 392
Query 65 VFFKVTQYPKITGAFLGSTSVELLAAAAREMQQAVPEELKLSPNADPDLFADPNFSVYEE 124
K+ +Y + F + E+ AD ++S
Sbjct 393 RKLKLCRYRMVEDEFRNPSVTEI-----------------------QKYLADEDYSG--- 426
Query 125 NGFLHLPWYAAALACRAAAAKLGRFPGSHPKPSAAAATAATGEAAAAATGGAAAARSVEE 184
+ +Y A AA +FPG GG ++E
Sbjct 427 ----AMGFYILLRAADRFAANYNKFPGQF-------------------DGG------MDE 457
Query 185 AIKQQQELVEKDSAAVLLELEEMEKAVGCR-LFAAAKVVQQLVSYRGSEFPATAALVGAV 243
I + K +A LL +GC ++ ++ + SE +A VG +
Sbjct 458 DISRL-----KTTALSLL------TDLGCNGSVLPDDLIHEMCRFGASEIHVVSAFVGGI 506
Query 244 AAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
A+QE IKL+ K+F P+ T+++NG+ + ++L
Sbjct 507 ASQEVIKLVTKQFVPMLGTYIFNGIDHKSQLLKL 540
> 7294767
Length=524
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 51/86 (59%), Gaps = 0/86 (0%)
Query 192 LVEKDSAAVLLELEEMEKAVGCRLFAAAKVVQQLVSYRGSEFPATAALVGAVAAQEAIKL 251
+VE+D + +M +G + V+ ++ Y G+E A +A +G AAQE IK+
Sbjct 439 IVEQDIGRLKSIAAKMLSDLGMHATISDDVLHEICRYGGAELHAVSAFIGGCAAQEVIKI 498
Query 252 LNKKFEPINNTFLWNGVQRRGLTMQL 277
+ K+++PI+NTF++N + +T++L
Sbjct 499 ITKQYKPIDNTFIYNAITTESVTLKL 524
Score = 30.8 bits (68), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 7/71 (9%)
Query 9 DFAADTKSYLELQKIYNEKAERD----ERRLRELLGQLDVHPSAKLNVSQEEIKCFCKNA 64
D A+T SY+ LQ IY ++A +D + +E L QL + + + + ++ CK A
Sbjct 324 DMTANTDSYIALQHIYRQQALQDADQVYHKCQEYLKQLALPADS---IDERSVRLICKEA 380
Query 65 VFFKVTQYPKI 75
V + +I
Sbjct 381 AGLAVIRGTRI 391
> SPAC323.06c
Length=500
Score = 53.1 bits (126), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 37/56 (66%), Gaps = 0/56 (0%)
Query 222 VQQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
+Q+L G E + ++ +G + AQE IKLL +++ P+NNTF+++GV R T +L
Sbjct 445 IQELERADGHELHSISSFIGGIVAQETIKLLAQQYLPLNNTFVFDGVHSRTETFKL 500
Score = 35.0 bits (79), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 36/72 (50%), Gaps = 9/72 (12%)
Query 6 LTADFAADTKSYLELQKIYNEKAERDERRLRELLGQLDVHPSAKLNVSQE-----EIKCF 60
L D T+ Y++LQ IY EK+E D + ++ + Q +LN S E EIK F
Sbjct 315 LLPDMNCSTQQYVKLQVIYKEKSENDILKFKKYVQQ----TLKRLNRSVEEITDLEIKHF 370
Query 61 CKNAVFFKVTQY 72
+N + KV +
Sbjct 371 SRNCLNIKVMDF 382
> CE01166
Length=525
Score = 52.8 bits (125), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 61/269 (22%), Positives = 104/269 (38%), Gaps = 53/269 (19%)
Query 9 DFAADTKSYLELQKIYNEKAERDERRLRELLGQLDVHPSAKLNVSQEEIKCFCKNAVFFK 68
D +D+ Y L +++EKA D + + L +++ +S + FCKNA +
Sbjct 310 DMTSDSSRYTRLATLFHEKALSDAQEVLRLTREVEKERGVGDVISDDVCYRFCKNADRIR 369
Query 69 VTQYPKITGAFLGSTSVELLAAAAREMQQAVPEELKLSPNADPDLFADPNFSVYEENGFL 128
V QY + E +A+ E+++ S N D + N V E
Sbjct 370 V-QYGDVLDY--------------NEETKAIVEKIRES-NIDEET---RNQKVDEAT--- 407
Query 129 HLPWYAAALACRAAAAKLGRFPGSHPKPSAAAATAATGEAAAAATGGAAAARSVEEAIKQ 188
W A + GR+PG++ P + A + EA+K
Sbjct 408 ---WMLLMRAVGRFQKEKGRYPGTNGVPVSIDAQDLKKRVEVL----------IREALKD 454
Query 189 QQELVEKDSAAVLLELEEMEKAVGCRLFAAAKVVQQLVSYRGSEFPATAALVGAVAAQEA 248
+Q+ + + E+ CR AA E ++ VG +AAQE
Sbjct 455 EQDFTSISNKVTDTAIAEI-----CRFGAA-------------ELHVISSYVGGIAAQEI 496
Query 249 IKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
IKL ++ PI+NTF+++G + T +
Sbjct 497 IKLATNQYVPIDNTFIFDGHTQESATFKF 525
> YPR180w
Length=347
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 222 VQQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGV 268
+QQ + +G EF AA++G AQ+ I +L K+ P+NN +++G+
Sbjct 292 IQQFIKQKGIEFAPVAAIIGGAVAQDVINILGKRLSPLNNFIVFDGI 338
> Hs4502169
Length=534
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/105 (29%), Positives = 49/105 (46%), Gaps = 7/105 (6%)
Query 180 RSVEEAIKQQQEL-------VEKDSAAVLLELEEMEKAVGCRLFAAAKVVQQLVSYRGSE 232
R+V+ KQQ VE+D + L + G + V + Y +E
Sbjct 430 RAVDRFHKQQGRYPGVSNYQVEEDIGKLKSCLTGFLQEYGLSVMVKDDYVHEFCRYGAAE 489
Query 233 FPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
AA +G AAQE IK++ K+F NNT++++G+ + T QL
Sbjct 490 PHTIAAFLGGAAAQEVIKIITKQFVIFNNTYIYSGMSQTSATFQL 534
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 39/72 (54%), Gaps = 7/72 (9%)
Query 4 RRLTADFAADTKSYLELQKIYNEKAERDE----RRLRELLGQLDVHPSAKLNVSQEEIKC 59
R D AD+ Y++LQ +Y EKA++D + +LL + P + +S++E+K
Sbjct 326 RGTIPDMIADSGKYIKLQNVYREKAKKDAAAVGNHVAKLLQSIGQAPES---ISEKELKL 382
Query 60 FCKNAVFFKVTQ 71
C N+ F +V +
Sbjct 383 LCSNSAFLRVVR 394
> At5g50680
Length=320
Score = 42.7 bits (99), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 35/57 (61%), Gaps = 1/57 (1%)
Query 221 VVQQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
++++LVS +EFP A++G + QE IK+++ K EP+ N F ++ +G+ L
Sbjct 261 LLERLVS-NNTEFPPACAIIGGILGQEVIKVISGKGEPLKNFFYFDAEDGKGVIEDL 316
> At5g50580
Length=320
Score = 42.7 bits (99), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 35/57 (61%), Gaps = 1/57 (1%)
Query 221 VVQQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
++++LVS +EFP A++G + QE IK+++ K EP+ N F ++ +G+ L
Sbjct 261 LLERLVS-NNTEFPPACAIIGGILGQEVIKVISGKGEPLKNFFYFDAEDGKGVIEDL 316
> YPL003w
Length=462
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 28/55 (50%), Gaps = 0/55 (0%)
Query 223 QQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
+ + R + + A G QEAIKL+ + PI+N FL+NG+ T ++
Sbjct 408 DEFIGLRVDDNYSVMAFFGGAVVQEAIKLITHHYVPIDNLFLYNGINNSSATYKI 462
> Hs4885585
Length=346
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 30/52 (57%), Gaps = 0/52 (0%)
Query 223 QQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLT 274
+ V Y SE A+VG + AQE +K L+++ P NN F ++G++ G+
Sbjct 289 EDFVRYCFSEMAPVCAVVGGILAQEIVKALSQRDPPHNNFFFFDGMKGNGIV 340
> CE05246
Length=343
Score = 38.9 bits (89), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 230 GSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGL 273
G F TAA VG V QEAIK +++ P+ N F++ G + G
Sbjct 293 GPNFGPTAACVGGVIGQEAIKSISEGKNPLRNLFIYTGFESTGF 336
> At4g24940
Length=319
Score = 36.2 bits (82), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 27/43 (62%), Gaps = 1/43 (2%)
Query 231 SEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGL 273
+EFP A+VG + AQ IK ++ K +P+ N F ++G +G+
Sbjct 268 TEFPPVCAIVGGILAQ-VIKAVSGKGDPLKNFFYYDGEDGKGV 309
> Hs20559018
Length=377
Score = 35.0 bits (79), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 30/50 (60%), Gaps = 2/50 (4%)
Query 224 QLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGL 273
+ ++Y SE A+VG + AQE +K L+++ P N F +NG++ G+
Sbjct 323 RFITYFFSEMAPVCAVVGGILAQEIVKALSQQDPPHN--FFFNGMKGNGI 370
> ECU04g0320
Length=607
Score = 30.4 bits (67), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 20/81 (24%), Positives = 38/81 (46%), Gaps = 0/81 (0%)
Query 190 QELVEKDSAAVLLELEEMEKAVGCRLFAAAKVVQQLVSYRGSEFPATAALVGAVAAQEAI 249
E V KD AVL L ++ + C ++VVQ ++ + G ++E I
Sbjct 318 DESVPKDRKAVLRLLGIEDRDLLCINLEPSEVVQYIIFRDRENGRVMVSFKGTTNSEETI 377
Query 250 KLLNKKFEPINNTFLWNGVQR 270
+ +N ++ ++ F+ NG +R
Sbjct 378 QDINCEYTEFSSGFVHNGFKR 398
> CE27034
Length=585
Score = 30.0 bits (66), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 7 TADFAADTKSYLELQKIYNEKAERDERRLRELLGQLDVHPSAKL 50
+ D A T+SY+ELQ +++ K +RD +L +L P A+L
Sbjct 515 SQDGADMTRSYMELQTMFSNKKKRDVSQLSSKDRRLKYEPIARL 558
> 7302229
Length=948
Score = 29.6 bits (65), Expect = 8.1, Method: Composition-based stats.
Identities = 28/103 (27%), Positives = 45/103 (43%), Gaps = 9/103 (8%)
Query 141 AAAAKLGRFPGSHPKPSAAAAT-AATGEAAAAATGGAAAARSVEEAIKQQQELVEKDSAA 199
+ +A G PG++P A T +G AAAAA+ +A V A Q Q +++
Sbjct 439 SQSASSGNNPGNNPAVQPNAPTPGQSGIAAAAASTNVVSATIVSSANVQGQSVIQPTPTI 498
Query 200 VLLELEEMEKAVGCRLFAAAKVVQQLVSYRGSEFPATAALVGA 242
+ + + L V+QQ ++ P AA+VGA
Sbjct 499 AFAAVAKHNTS----LLENGPVLQQQLAVT----PTVAAIVGA 533
Lambda K H
0.316 0.129 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6021067820
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40