bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2523_orf2
Length=132
Score E
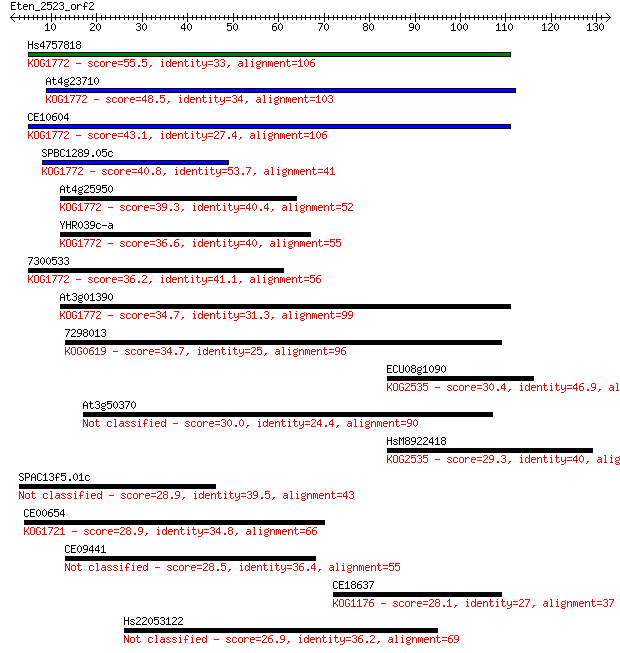
Sequences producing significant alignments: (Bits) Value
Hs4757818 55.5 3e-08
At4g23710 48.5 3e-06
CE10604 43.1 1e-04
SPBC1289.05c 40.8 7e-04
At4g25950 39.3 0.002
YHR039c-a 36.6 0.012
7300533 36.2 0.016
At3g01390 34.7 0.054
7298013 34.7 0.055
ECU08g1090 30.4 0.94
At3g50370 30.0 1.4
HsM8922418 29.3 1.9
SPAC13f5.01c 28.9 2.8
CE00654 28.9 2.9
CE09441 28.5 3.7
CE18637 28.1 4.8
Hs22053122 26.9 9.9
> Hs4757818
Length=118
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 35/106 (33%), Positives = 62/106 (58%), Gaps = 0/106 (0%)
Query 5 MKTSNALIQQLLRAEEEAEQIVHKARENRVKMLKDARASAEEELRAFRLKEEERFKVEVE 64
M + + IQQLL+AE+ A + V +AR+ + + LK A+ A+ E+ +RL+ E+ FK +
Sbjct 1 MASQSQGIQQLLQAEKRAAEKVSEARKRKNRRLKQAKEEAQAEIEQYRLQREKEFKAKEA 60
Query 65 QRLGQDDSLTNELADRTKAEIEIIKKDYVANKDGVLDFISRKVLDV 110
LG S + E+ T+ ++ I++ + N+D VLD + V D+
Sbjct 61 AALGSRGSCSTEVEKETQEKMTILQTYFRQNRDEVLDNLLAFVCDI 106
> At4g23710
Length=106
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 55/103 (53%), Gaps = 0/103 (0%)
Query 9 NALIQQLLRAEEEAEQIVHKARENRVKMLKDARASAEEELRAFRLKEEERFKVEVEQRLG 68
+A IQQLL AE EA+QIV+ AR ++ LK A+ AE E+ + E+ F+ ++E G
Sbjct 3 SAGIQQLLAAEREAQQIVNAARTAKMTRLKQAKEEAETEVAEHKTSTEQGFQRKLEATSG 62
Query 69 QDDSLTNELADRTKAEIEIIKKDYVANKDGVLDFISRKVLDVD 111
+ L T A+IE +K + V+D + + V V+
Sbjct 63 DSGANVKRLEQETDAKIEQLKNEATRISKDVVDMLLKNVTTVN 105
> CE10604
Length=126
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 59/106 (55%), Gaps = 0/106 (0%)
Query 5 MKTSNALIQQLLRAEEEAEQIVHKARENRVKMLKDARASAEEELRAFRLKEEERFKVEVE 64
M + IQQLL AE+ A + +++AR+ +++ K A+ A+ E+ ++ + E FK +
Sbjct 1 MASQTQGIQQLLAAEKRAAEKINEARKRKLQRTKQAKQEAQAEVEKYKQQREAEFKAFEQ 60
Query 65 QRLGQDDSLTNELADRTKAEIEIIKKDYVANKDGVLDFISRKVLDV 110
Q LG + + +++ T+ +I +K+ NK V+ + + V D+
Sbjct 61 QYLGTKEDIESKIRRDTEDQISGMKQSVAGNKQAVIVRLLQLVCDI 106
> SPBC1289.05c
Length=108
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 22/41 (53%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 8 SNALIQQLLRAEEEAEQIVHKARENRVKMLKDARASAEEEL 48
+N+ IQQLL AE+ A IV KAR++R + LKDAR A+ E+
Sbjct 5 TNSGIQQLLEAEKVARNIVEKARQHRTQRLKDARLEAKREI 45
> At4g25950
Length=108
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 34/52 (65%), Gaps = 0/52 (0%)
Query 12 IQQLLRAEEEAEQIVHKARENRVKMLKDARASAEEELRAFRLKEEERFKVEV 63
IQ LL AE+EA +IV AR ++ +K A+ AE+E+ +R + EE ++ +V
Sbjct 10 IQMLLTAEQEAGRIVSAARTAKLARMKQAKDEAEKEMEEYRSRLEEEYQTQV 61
> YHR039c-a
Length=114
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 38/55 (69%), Gaps = 1/55 (1%)
Query 12 IQQLLRAEEEAEQIVHKARENRVKMLKDARASAEEELRAFRLKEEERFKVEVEQR 66
I LL+AE+EA +IV KAR+ R LK A+ A +E+ ++++++++ K E EQ+
Sbjct 7 IATLLQAEKEAHEIVSKARKYRQDKLKQAKTDAAKEIDSYKIQKDKELK-EFEQK 60
> 7300533
Length=117
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 34/56 (60%), Gaps = 0/56 (0%)
Query 5 MKTSNALIQQLLRAEEEAEQIVHKARENRVKMLKDARASAEEELRAFRLKEEERFK 60
M + IQQLL AE++A + V +AR+ + + LK A+ A EE+ FR + E FK
Sbjct 1 MASQTQGIQQLLAAEKKAAEKVAEARKRKARRLKQAKDEATEEIEKFRQERERAFK 56
> At3g01390
Length=110
Score = 34.7 bits (78), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 31/99 (31%), Positives = 54/99 (54%), Gaps = 0/99 (0%)
Query 12 IQQLLRAEEEAEQIVHKARENRVKMLKDARASAEEELRAFRLKEEERFKVEVEQRLGQDD 71
IQQLL AE EA+ IV+ AR ++ LK A+ AE+E+ ++ + E+ F+ ++E+ G
Sbjct 10 IQQLLAAEVEAQHIVNAARTAKMARLKQAKEEAEKEIAEYKAQTEQDFQRKLEETSGDSG 69
Query 72 SLTNELADRTKAEIEIIKKDYVANKDGVLDFISRKVLDV 110
+ L T +IE +K + V++ + + V V
Sbjct 70 ANVKRLEQETDTKIEQLKNEASRISKDVVEMLLKHVTTV 108
> 7298013
Length=1257
Score = 34.7 bits (78), Expect = 0.055, Method: Composition-based stats.
Identities = 24/99 (24%), Positives = 47/99 (47%), Gaps = 7/99 (7%)
Query 13 QQLLRAEEEAEQIVHKAREN---RVKMLKDARASAEEELRAFRLKEEERFKVEVEQRLGQ 69
QQL R E Q VH+ REN +V+ ++ + S + +R FR +Q Q
Sbjct 843 QQLERLRENYTQQVHRIRENCAQQVEWIQSSYTSQAKHIREFRDIGSNHLTTLKDQYYDQ 902
Query 70 DDSLTNELADRTKAEIEIIKKDYVANKDGVLDFISRKVL 108
++ D + ++ ++++YV ++ + F + +VL
Sbjct 903 ----VKKVRDYSTGQLSWVRENYVFQRNKIRKFSAHQVL 937
> ECU08g1090
Length=613
Score = 30.4 bits (67), Expect = 0.94, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 24/32 (75%), Gaps = 1/32 (3%)
Query 84 EIEIIKKDYVANKDGVLDFISRKVLDVDIVIG 115
E+E++++DYVANK G F+S + + DI+IG
Sbjct 460 EVELVRRDYVANK-GWETFLSYEDVKQDILIG 490
> At3g50370
Length=2152
Score = 30.0 bits (66), Expect = 1.4, Method: Composition-based stats.
Identities = 22/93 (23%), Positives = 41/93 (44%), Gaps = 7/93 (7%)
Query 17 RAEEEAEQIVHKARENRVKMLKDA---RASAEEELRAFRLKEEERFKVEVEQRLGQDDSL 73
R EEEA + + + R++ + A R S EEE ++EE R + ++ L ++ +
Sbjct 518 RLEEEAREAAFRNEQERLEATRRAEELRKSKEEEKHRLFMEEERRKQAAKQKLLELEEKI 577
Query 74 TNELADRTKAEIEIIKKDYVANKDGVLDFISRK 106
+ A+ K ++D LD + K
Sbjct 578 SRRQAEAAKG----CSSSSTISEDKFLDIVKEK 606
> HsM8922418
Length=499
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 29/45 (64%), Gaps = 4/45 (8%)
Query 84 EIEIIKKDYVANKDGVLDFISRKVLDVDIVIGAKKVAILRSYAER 128
++E++++DYVAN G F+S + D DI+IG + LR +ER
Sbjct 419 QVELVRRDYVAN-GGWETFLSYEDPDQDILIGLLR---LRKCSER 459
> SPAC13f5.01c
Length=780
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 23/43 (53%), Gaps = 4/43 (9%)
Query 3 NNMKTSNALIQQLLRAEEEAEQIVHKARENRVKMLKDARASAE 45
NN NAL++Q + EEE E I +A E +L+D A E
Sbjct 328 NNAVDENALMKQKINEEEETEVIAQEAEE----ILQDENAQVE 366
> CE00654
Length=124
Score = 28.9 bits (63), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 35/69 (50%), Gaps = 11/69 (15%)
Query 4 NMKTSNALIQQ---LLRAEEEAEQIVHKARENRVKMLKDARASAEEELRAFRLKEEERFK 60
N KTS L++ RA+ E V+K ++ K L + S E FRLKE+ +
Sbjct 36 NFKTSEELVEHKKNYPRADVEKALSVYKEKDTIYKCL---QCSGE-----FRLKEDFDYH 87
Query 61 VEVEQRLGQ 69
E+ +RLG+
Sbjct 88 QELHKRLGE 96
> CE09441
Length=1229
Score = 28.5 bits (62), Expect = 3.7, Method: Composition-based stats.
Identities = 20/59 (33%), Positives = 31/59 (52%), Gaps = 4/59 (6%)
Query 13 QQLLRAEEEAEQIVHKAR----ENRVKMLKDARASAEEELRAFRLKEEERFKVEVEQRL 67
++LL + + ++I KAR E + KML+ E E R L E+ER + + QRL
Sbjct 1091 EELLEKQLQEQEIEEKARNEMIERKQKMLQQLEELKEAEERQRVLLEQERLQEQERQRL 1149
> CE18637
Length=550
Score = 28.1 bits (61), Expect = 4.8, Method: Composition-based stats.
Identities = 10/37 (27%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 72 SLTNELADRTKAEIEIIKKDYVANKDGVLDFISRKVL 108
++ NEL+ ++KK +VA +D + D+++++V+
Sbjct 455 AINNELSGERPVAFVVLKKGFVATEDDLKDYVNKRVI 491
> Hs22053122
Length=412
Score = 26.9 bits (58), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 40/81 (49%), Gaps = 14/81 (17%)
Query 26 VHKARENRVKMLKDARASAEEELRAFRLKEEER-----------FKVEVEQRLGQDDS-L 73
+H R + ML++ S EE +A R EE R F E + LG+++S
Sbjct 121 LHHKRLAKECMLQEENKSLREENKALR--EENRMLSKENKILQVFWEEHKASLGREESRA 178
Query 74 TNELADRTKAEIEIIKKDYVA 94
+ L + A +E++KKD+VA
Sbjct 179 PSPLLHKDSASLEVVKKDHVA 199
Lambda K H
0.313 0.130 0.329
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1319765976
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40