bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
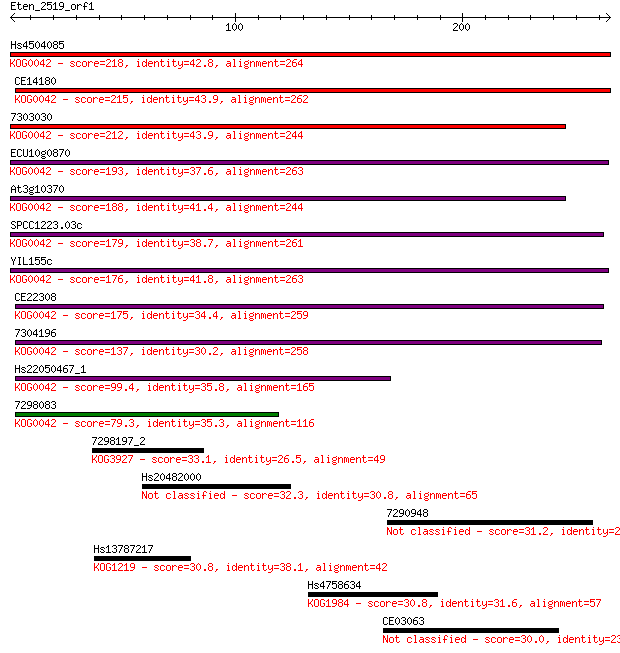
Query= Eten_2519_orf1
Length=264
Score E
Sequences producing significant alignments: (Bits) Value
Hs4504085 218 1e-56
CE14180 215 8e-56
7303030 212 6e-55
ECU10g0870 193 3e-49
At3g10370 188 8e-48
SPCC1223.03c 179 7e-45
YIL155c 176 3e-44
CE22308 175 8e-44
7304196 137 3e-32
Hs22050467_1 99.4 7e-21
7298083 79.3 8e-15
7298197_2 33.1 0.69
Hs20482000 32.3 1.00
7290948 31.2 2.3
Hs13787217 30.8 3.0
Hs4758634 30.8 3.3
CE03063 30.0 5.7
> Hs4504085
Length=727
Score = 218 bits (554), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 113/269 (42%), Positives = 167/269 (62%), Gaps = 18/269 (6%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQ-----IIA 55
+ +Y+DGQ ND+RM L++AL+ G AA AN++ +K + Q +
Sbjct 215 AIVYYDGQHNDARMNLAIALTAARYG-------AATANYMEVVSLLKKTDPQTGKVHVSG 267
Query 56 VKVQDRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKT 115
+ +D T +EF + K V+N TGP TDSVRKMD +DAA++ P+AG HIV+P +Y+ ++
Sbjct 268 ARCKDVLTGQEFDVRAKCVINATGPFTDSVRKMDDKDAAAICQPSAGVHIVMPGYYSPES 327
Query 116 PFGLLLPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMD 175
GLL P TSDGRV+F LPW+ T+AGTTD P + P P +++F++NE+ YL D
Sbjct 328 -MGLLDPATSDGRVIFFLPWQKMTIAGTTDTPTDVTHHPIPSEEDINFILNEVRNYLSCD 386
Query 176 PQQMRGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMA 235
+ RGD+ + W+G RPL++ A DT + R+H V + ESGLI++ GGKWTTYR MA
Sbjct 387 VEVRRGDVLAAWSGIRPLVTDPKSA-DTQSISRNHVVDIS-ESGLITIAGGKWTTYRSMA 444
Query 236 QDAVDTLLEVHKDKVAASMPCRTKGMKVQ 264
+D ++ ++ H K P RT G+ +Q
Sbjct 445 EDTINAAVKTHNLKAG---PSRTVGLFLQ 470
> CE14180
Length=722
Score = 215 bits (547), Expect = 8e-56, Method: Compositional matrix adjust.
Identities = 115/263 (43%), Positives = 165/263 (62%), Gaps = 13/263 (4%)
Query 3 LYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQDRE 62
+Y+DGQ ND+RM L++ L+ G AA ANHV ++ K+E G++I V+D
Sbjct 222 IYYDGQHNDARMNLAIILTAIRHG-------AACANHVRVEKLNKDETGKVIGAHVRDMV 274
Query 63 TDEEFTIHCKVVVNCTGPLTDSVRKM-DHEDAASLVIPAAGTHIVLPHWYTHKTPFGLLL 121
T E+ I K V+N TGP TDS+R M D E A + P++G HI LP +Y+ + GLL
Sbjct 275 TGGEWDIKAKAVINATGPFTDSIRLMGDPETARPICAPSSGVHITLPGYYS-PSNTGLLD 333
Query 122 PQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQMRG 181
P TSDGRV+F LPWE T+AGTTDAP++ P+P +++F++ E+ YL D RG
Sbjct 334 PDTSDGRVIFFLPWERMTIAGTTDAPSDVTLSPQPTDHDIEFILQEIRGYLSKDVSVRRG 393
Query 182 DIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQDAVDT 241
D+ S W+G RPL+ ++ KDT + R+H + V +SGLI++ GGKWTTYR MA++ VD
Sbjct 394 DVMSAWSGLRPLVRDPNK-KDTKSLARNHIIEVG-KSGLITIAGGKWTTYRHMAEETVDR 451
Query 242 LLEVHKDKVAASMPCRTKGMKVQ 264
++EVH K C T G+ ++
Sbjct 452 VVEVHGLKTENG--CVTPGLLLE 472
> 7303030
Length=700
Score = 212 bits (539), Expect = 6e-55, Method: Compositional matrix adjust.
Identities = 107/248 (43%), Positives = 153/248 (61%), Gaps = 14/248 (5%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQ----IIAV 56
+ +Y+DGQ +D+RMCL++AL+ G A NHV KE +K ++G +
Sbjct 209 AIVYYDGQQDDARMCLAVALTAARHG-------ATVCNHVEVKELLKKDDGTGKQVLCGA 261
Query 57 KVQDRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTP 116
KV+D + +EFT+ K +VN GP TDS+RKMD+ S+ P++G HIVLP +Y+
Sbjct 262 KVKDHISGKEFTVKAKCIVNAAGPFTDSIRKMDNPTVKSICCPSSGVHIVLPGYYS-PDQ 320
Query 117 FGLLLPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDP 176
GLL P TSDGRV+F LPW+ +T+AGTTD P P P E+ F++NE+ YL D
Sbjct 321 MGLLDPSTSDGRVIFFLPWQRQTIAGTTDLPCEITHNPTPTEDEIQFILNEIKNYLNADV 380
Query 177 QQMRGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQ 236
+ RGD+ S W+G RPL+S ++ DT + R+H V V S LI++ GGKWTTYR MA+
Sbjct 381 EVRRGDVLSAWSGIRPLVSDPNK-DDTQSLARNHIVHVS-PSNLITIAGGKWTTYRAMAE 438
Query 237 DAVDTLLE 244
+D ++
Sbjct 439 HTIDAAIK 446
> ECU10g0870
Length=614
Score = 193 bits (491), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 99/263 (37%), Positives = 157/263 (59%), Gaps = 13/263 (4%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQD 60
+ +YFDGQ +D+R + + ++ G A AANHV+A+ + E G+I+ V+ +D
Sbjct 201 AMVYFDGQQDDARNNVMIVMTAVCHG-------AVAANHVSARSLMI-EGGKIVGVRCRD 252
Query 61 RETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGLL 120
T E I V+N TG L D +R+MD DA +++ ++GTHIV+P Y K G L
Sbjct 253 EITGSEIEIRGTGVINSTGNLADDLRRMDDADAREIIVQSSGTHIVIPKEYAPK-EMGFL 311
Query 121 LPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQMR 180
P TSD R+ F +PW G+T+ G+TD P P +++FL++E+ AY M P+ R
Sbjct 312 DPLTSDNRIAFFMPWMGKTIVGSTDIKTKTELSPSPTEEDLEFLIHEVQAYTSMHPKLTR 371
Query 181 GDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQDAVD 240
++ +VW G RPL+ + + DT +VR H V ++ ++GL+++ GGKWT YR+MA+DA+D
Sbjct 372 DEVSAVWTGIRPLV-KDPDVSDTGSIVRKHFVRIE-KNGLLTVTGGKWTIYRKMAEDAID 429
Query 241 TLLEVHKDKVAASMPCRTKGMKV 263
L + + S PC TK +++
Sbjct 430 --LAISAFSLKPSGPCVTKYVRI 450
> At3g10370
Length=629
Score = 188 bits (478), Expect = 8e-48, Method: Compositional matrix adjust.
Identities = 101/245 (41%), Positives = 151/245 (61%), Gaps = 13/245 (5%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNE-NGQIIAVKVQ 59
+ +Y+DGQMNDSR+ + LA + + AA NH I ++ +II +++
Sbjct 223 TVVYYDGQMNDSRLNVGLACTAA-------LAGAAVLNHAEVVSLITDDATKRIIGARIR 275
Query 60 DRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGL 119
+ T +EF + KVVVN GP DS+RKM ED ++ P++G HIVLP +Y+ + GL
Sbjct 276 NNLTGQEFNSYAKVVVNAAGPFCDSIRKMIDEDTKPMICPSSGVHIVLPDYYSPEG-MGL 334
Query 120 LLPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQM 179
++P+T DGRV+F+LPW GRTVAGTTD+ + + P P E+ F+++ +S YL + +
Sbjct 335 IVPKTKDGRVVFMLPWLGRTVAGTTDSNTSITSLPEPHEDEIQFILDAISDYLNIKVR-- 392
Query 180 RGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQDAV 239
R D+ S W+G RPL + AK T + R H V + GL+++ GGKWTTYR MA+DAV
Sbjct 393 RTDVLSAWSGIRPL-AMDPTAKSTESISRDHVVF-EENPGLVTITGGKWTTYRSMAEDAV 450
Query 240 DTLLE 244
D ++
Sbjct 451 DAAIK 455
> SPCC1223.03c
Length=649
Score = 179 bits (453), Expect = 7e-45, Method: Compositional matrix adjust.
Identities = 101/279 (36%), Positives = 153/279 (54%), Gaps = 30/279 (10%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQD 60
S +Y DG ND+RM +LA++ G A N++ K+ +K+++ ++ V D
Sbjct 213 SCVYHDGSFNDTRMNTTLAVTAIDNG-------ATVLNYMEVKKLLKSKDNKLEGVLAID 265
Query 61 RETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAAS---------LVIPAAGTHIVLPHWY 111
RET +E+ I VVN TGP +D + +MD + +V+P+AG H+VLP +Y
Sbjct 266 RETGKEYQIKATSVVNATGPFSDKILEMDADPQGEPPKTAQFPRMVVPSAGVHVVLPEYY 325
Query 112 THKTPFGLLLPQTSDGRVLFLLPWEGRTVAGTTDAPANKA-AEPRPGVSEVDFLVNELSA 170
G+L P TSD RV+F LPW+G+ +AGTTD P + P P ++ ++ EL
Sbjct 326 CPPN-IGILDPSTSDNRVMFFLPWQGKVIAGTTDKPLSSVPTNPTPSEDDIQLILKELQK 384
Query 171 YLRMDPQQMRGDIQSVWAGHRPLISQTSEAK--------DTAGVVRSHTVLVDHESGLIS 222
YL R D+ S W G RPL+ S +T G+VRSH + ++GL++
Sbjct 385 YLVFPVD--REDVLSAWCGIRPLVRDPSTVPPGTDPTTGETQGLVRSHFIF-KSDTGLLT 441
Query 223 LMGGKWTTYRRMAQDAVDTLLEVHKDKVAASMPCRTKGM 261
+ GGKWTTYR MA++ V+ L++ H D A PC+TK +
Sbjct 442 ISGGKWTTYREMAEETVNELIKDH-DFGKALKPCQTKKL 479
> YIL155c
Length=649
Score = 176 bits (447), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 110/299 (36%), Positives = 163/299 (54%), Gaps = 52/299 (17%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKN-ENGQIIAVKVQ 59
S +Y DG NDSR+ +LA++ G A N+V ++ IK+ +G++I + +
Sbjct 213 SLVYHDGSFNDSRLNATLAITAVENG-------ATVLNYVEVQKLIKDPTSGKVIGAEAR 265
Query 60 DRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAA------------------------S 95
D ET+E I+ K VVN TGP +D++ +MD +
Sbjct 266 DVETNELVRINAKCVVNATGPYSDAILQMDRNPSGLPDSPLNDNSKIKSTFNQIAVMDPK 325
Query 96 LVIPAAGTHIVLPHWYTHKTPFGLLLPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAE-P 154
+VIP+ G HIVLP +Y K GLL +TSDGRV+F LPW+G+ +AGTTD P + E P
Sbjct 326 MVIPSIGVHIVLPSFYCPKD-MGLLDVRTSDGRVMFFLPWQGKVLAGTTDIPLKQVPENP 384
Query 155 RPGVSEVDFLVNELSAYLRMDPQQMRGDIQSVWAGHRPLI-------SQTSEAKDTAGVV 207
P +++ ++ EL Y+ + R D+ S WAG RPL+ + + T GVV
Sbjct 385 MPTEADIQDILKELQHYIEFPVK--REDVLSAWAGVRPLVRDPRTIPADGKKGSATQGVV 442
Query 208 RSHTVLVDHESGLISLMGGKWTTYRRMAQDAVDTLLEV---HKDKVAASMPCRTKGMKV 263
RSH L ++GLI++ GGKWTTYR+MA++ VD ++EV H K PC T+ +K+
Sbjct 443 RSH-FLFTSDNGLITIAGGKWTTYRQMAEETVDKVVEVGGFHNLK-----PCHTRDIKL 495
> CE22308
Length=673
Score = 175 bits (444), Expect = 8e-44, Method: Compositional matrix adjust.
Identities = 89/259 (34%), Positives = 146/259 (56%), Gaps = 11/259 (4%)
Query 3 LYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQDRE 62
LY+DGQ ND+R+ L +AL+ G A NH +K+ G++ V+D
Sbjct 210 LYYDGQGNDARLVLVVALTAIRNG-------AKCVNHTECIGLLKDSYGKVNGALVKDHI 262
Query 63 TDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGLLLP 122
+ E + IH KVVVN TGP D +R+M E +++ ++G H+ + ++ GL++P
Sbjct 263 SGETYEIHSKVVVNATGPFNDHIREMADETRNKIIVGSSGIHLTVAKYFCPGNA-GLIVP 321
Query 123 QTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQMRGD 182
++SDGRV+F PWE T+ GTTD P + P E+ +++ E++ + + R D
Sbjct 322 KSSDGRVIFAFPWENVTIVGTTDDPTEPSHSPTVTEKEIQYILKEMNRAFEKEYEIKRED 381
Query 183 IQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQDAVDTL 242
+ S+W+G R L+ K+ + R H V V +GLI++ GGK TT+R MA++ V+ L
Sbjct 382 VTSIWSGIRGLVQDPKRQKEHNSLARGHLVDVG-PTGLITIAGGKLTTFRHMAEETVNKL 440
Query 243 LEVHKDKVAASMPCRTKGM 261
L++ +K+ + PC T+GM
Sbjct 441 LDI--NKILEAQPCVTRGM 457
> 7304196
Length=713
Score = 137 bits (344), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 78/265 (29%), Positives = 147/265 (55%), Gaps = 19/265 (7%)
Query 3 LYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQDRE 62
++++GQ +D+RMCLS+AL+ G A NH+ IKN++G + + D+
Sbjct 201 VFYEGQHDDARMCLSVALTAARYG-------ADICNHMKVSRLIKNKDGNVAGAEAVDQL 253
Query 63 TDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGLLLP 122
T ++++I KVVVN TG ++DS+R+M+ +A L + + IV+P +Y + G+ P
Sbjct 254 TGKKYSIRAKVVVNATGHMSDSLRQMEDSEAKPLCTRSWSSIIVMPRFYCPEE-VGVFDP 312
Query 123 QTSDGRVLFLLPWEGRTVAGTTDAPAN-KAAEPRPGVSEVDFLVNELSAYLRMDPQQMRG 181
T GR +F LPW+G T+ G++D P N + +P ++V L++ + ++ + R
Sbjct 313 HTPSGRSIFFLPWQGHTLVGSSDEPLNLPEQDTQPTETDVQSLLDCVKHFVTPNFDVRRC 372
Query 182 DIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQDAVDT 241
D+ +VW G++PL Q+ T ++H + + + ++S++GG W+++R +A+ V+
Sbjct 373 DVLAVWGGYKPLPVQSDTLHHTR--FKNHVIQLGPRN-MLSIVGGTWSSFRVLAEKTVNA 429
Query 242 LLE------VHKDKVAASMPCRTKG 260
+ + K+ V A C+ G
Sbjct 430 AIRYGDLSPLRKESVTAE-SCKLDG 453
> Hs22050467_1
Length=375
Score = 99.4 bits (246), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 59/166 (35%), Positives = 88/166 (53%), Gaps = 21/166 (12%)
Query 3 LYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQDRE 62
+Y+DG+ ND+ M L++ L+ G A N +K + Q D E
Sbjct 91 VYYDGRHNDAWMNLAIVLTAARYG-------DATVNDREVVSLLKKTDTQ------TDSE 137
Query 63 TD-EEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGLLL 121
+ +EF + K V+N G TDSV K+D +DAA++ P AG HIV+P + + L
Sbjct 138 SAWQEFDMKAKCVINAVGHFTDSVCKIDDKDAAAICQPTAGVHIVMPA----QRAWDFL- 192
Query 122 PQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNE 167
TSDGRV+F LPW+ +AGTTD P + P P +++F++NE
Sbjct 193 --TSDGRVIFFLPWQKMMIAGTTDTPTDITHHPSPSEEDINFILNE 236
> 7298083
Length=330
Score = 79.3 bits (194), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 41/118 (34%), Positives = 66/118 (55%), Gaps = 10/118 (8%)
Query 3 LYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQDRE 62
+Y+DGQ++D+RMC++L ++ G A NH+ E + E G V V+D+
Sbjct 212 VYYDGQVDDARMCVALVMTAVALG-------ANVGNHMEVVEIMPQE-GCCRVVSVKDKI 263
Query 63 TDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWY--THKTPFG 118
+ E+F I K V+N TG TD++R+MD E A +++P T + LP ++ H P G
Sbjct 264 SCEQFYIQSKAVINATGSSTDAIRQMDKEGTAPILLPTLETQVSLPRYFGSGHYEPHG 321
> 7298197_2
Length=302
Score = 33.1 bits (74), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 37 ANHVAAKEFIKNENGQIIAVKVQDRETDEEFTIHCKVVVNCTGPLTDSV 85
AN F N+ +I+A+K+++ + +E I+CK+V+ + P+ +++
Sbjct 226 ANEGKKSFFGPNDVNRIVALKIKNLKANERVHINCKIVIQSSHPVCNTI 274
> Hs20482000
Length=328
Score = 32.3 bits (72), Expect = 1.00, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 29/65 (44%), Gaps = 4/65 (6%)
Query 59 QDRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFG 118
R T + ++ VV+ PL S+R D +DA +I HI HWY PF
Sbjct 268 NSRHTVKVASVFYTVVIPLLNPLIYSLRNKDVKDAIRKIINTKYFHIKHRHWY----PFN 323
Query 119 LLLPQ 123
++ Q
Sbjct 324 FVIEQ 328
> 7290948
Length=1022
Score = 31.2 bits (69), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 24/90 (26%), Positives = 42/90 (46%), Gaps = 6/90 (6%)
Query 167 ELSAYLRMDPQQMRGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGG 226
+LS +R+ ++ G + +V R + QT + K + + R T HE G + G
Sbjct 350 DLSLQIRVLAAELLGGMTAV---SREFLHQTLDKKLMSNLRRKRTA---HERGARLVASG 403
Query 227 KWTTYRRMAQDAVDTLLEVHKDKVAASMPC 256
+W++ +R A DA L+ + AS C
Sbjct 404 EWSSGKRWADDAPQEHLDAQSISIIASGAC 433
> Hs13787217
Length=4349
Score = 30.8 bits (68), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 38 NHVAAKEFIKNENGQIIAVKVQDRETDEEFTIHCKVVVNCTG 79
N +A+++F N NGQI ++ DRE E I KV+ G
Sbjct 2522 NKLASEKFSINPNGQIATLQKLDRENSTERVIAIKVMARDGG 2563
> Hs4758634
Length=1125
Score = 30.8 bits (68), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 132 LLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQMRGDIQSVWA 188
LLP V TT+ PA +A+E R ++ L N L+ +L + +G +QS+++
Sbjct 951 LLPLTKSPVESTTEPPAVRASEERLSNGDIYLLENGLNLFLWVGASVQQGVVQSLFS 1007
> CE03063
Length=731
Score = 30.0 bits (66), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 18/77 (23%), Positives = 31/77 (40%), Gaps = 7/77 (9%)
Query 165 VNELSAYLRMDPQQMRGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLM 224
+NE+ Y QM ++ HRPL+ RSH++ D++S
Sbjct 314 INEIEKYTESPAVQMLDKPRAGGRAHRPLLQD-------VIFTRSHSITSDYDSNKEHFD 366
Query 225 GGKWTTYRRMAQDAVDT 241
G W++ + +D T
Sbjct 367 SGSWSSTQSRYKDGRST 383
Lambda K H
0.318 0.132 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5613892628
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40