bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2501_orf2
Length=157
Score E
Sequences producing significant alignments: (Bits) Value
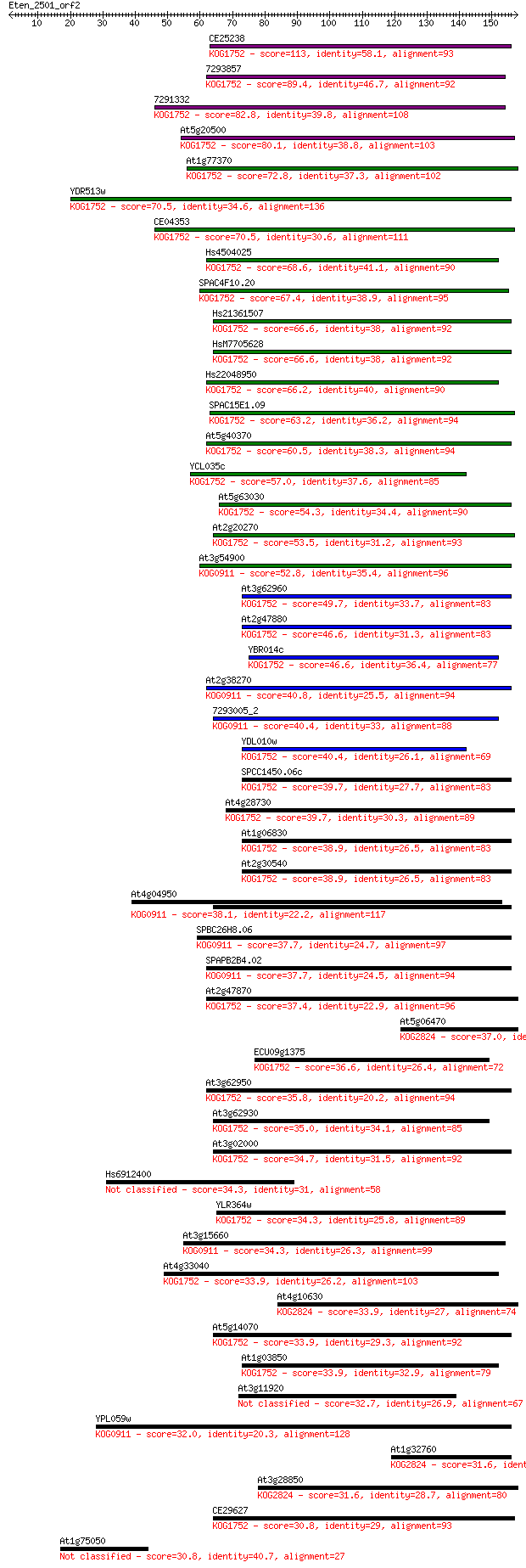
CE25238 113 2e-25
7293857 89.4 3e-18
7291332 82.8 3e-16
At5g20500 80.1 2e-15
At1g77370 72.8 2e-13
YDR513w 70.5 1e-12
CE04353 70.5 1e-12
Hs4504025 68.6 5e-12
SPAC4F10.20 67.4 1e-11
Hs21361507 66.6 2e-11
HsM7705628 66.6 2e-11
Hs22048950 66.2 2e-11
SPAC15E1.09 63.2 2e-10
At5g40370 60.5 1e-09
YCL035c 57.0 1e-08
At5g63030 54.3 1e-07
At2g20270 53.5 2e-07
At3g54900 52.8 3e-07
At3g62960 49.7 2e-06
At2g47880 46.6 2e-05
YBR014c 46.6 2e-05
At2g38270 40.8 0.001
7293005_2 40.4 0.001
YDL010w 40.4 0.001
SPCC1450.06c 39.7 0.003
At4g28730 39.7 0.003
At1g06830 38.9 0.004
At2g30540 38.9 0.004
At4g04950 38.1 0.007
SPBC26H8.06 37.7 0.008
SPAPB2B4.02 37.7 0.009
At2g47870 37.4 0.013
At5g06470 37.0 0.016
ECU09g1375 36.6 0.021
At3g62950 35.8 0.032
At3g62930 35.0 0.058
At3g02000 34.7 0.075
Hs6912400 34.3 0.098
YLR364w 34.3 0.10
At3g15660 34.3 0.11
At4g33040 33.9 0.12
At4g10630 33.9 0.12
At5g14070 33.9 0.14
At1g03850 33.9 0.14
At3g11920 32.7 0.31
YPL059w 32.0 0.51
At1g32760 31.6 0.61
At3g28850 31.6 0.68
CE29627 30.8 1.1
At1g75050 30.8 1.1
> CE25238
Length=105
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 54/95 (56%), Positives = 66/95 (69%), Gaps = 2/95 (2%)
Query 63 WVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVE--QIENNPHCNAIQDYLKEK 120
+VD L+ KV +F+K++CPYC A AAL S+NV ++ +I+ CN IQDYL
Sbjct 5 FVDGLLQSSKVVVFSKSYCPYCHKARAALESVNVKPDALQWIEIDERKDCNEIQDYLGSL 64
Query 121 TGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
TGARSVPRVFING FFGGGDDT AG KNG L L+
Sbjct 65 TGARSVPRVFINGKFFGGGDDTAAGAKNGKLAALL 99
> 7293857
Length=114
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 43/92 (46%), Positives = 59/92 (64%), Gaps = 1/92 (1%)
Query 62 QWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKT 121
++V++ + +KV IF+KT+CPYC A + LNV D + +++ NP N IQ L E T
Sbjct 21 KFVENTIASNKVVIFSKTYCPYCTMAKEPFKKLNV-DATIIELDGNPDGNEIQAVLGEIT 79
Query 122 GARSVPRVFINGTFFGGGDDTVAGVKNGTLQK 153
GAR+VPRVFI+G F GGG D + G LQK
Sbjct 80 GARTVPRVFIDGKFIGGGTDIKRMFETGALQK 111
> 7291332
Length=116
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 43/108 (39%), Positives = 61/108 (56%), Gaps = 1/108 (0%)
Query 46 TLFASTMALNSAQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIE 105
TL T+ ++ Q+V ++G+KV IF+K++CPYC A R +NV +E ++
Sbjct 7 TLQRPTLYVSMDSSHAQFVRDTISGNKVVIFSKSYCPYCSMAKEQFRKINVKATVIE-LD 65
Query 106 NNPHCNAIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQK 153
N IQ L E TG+R+VPR FI+G F GGG D + G LQK
Sbjct 66 QRDDGNEIQAVLGEMTGSRTVPRCFIDGKFVGGGTDVKRLYEQGILQK 113
> At5g20500
Length=135
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 40/105 (38%), Positives = 63/105 (60%), Gaps = 3/105 (2%)
Query 54 LNSAQDVPQ--WVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCN 111
++SA P+ +V ++ HK+ IF+K++CPYC+ A + R L+ +V +++
Sbjct 23 VSSAASSPEADFVKKTISSHKIVIFSKSYCPYCKKAKSVFRELDQVP-YVVELDEREDGW 81
Query 112 AIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
+IQ L E G R+VP+VFING GG DDTV ++G L KL+
Sbjct 82 SIQTALGEIVGRRTVPQVFINGKHLGGSDDTVDAYESGELAKLLG 126
> At1g77370
Length=130
Score = 72.8 bits (177), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 58/102 (56%), Gaps = 1/102 (0%)
Query 56 SAQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQD 115
+A V +V + + +K+ IF+K++CPYC + L VE ++ + IQ
Sbjct 29 AANSVSAFVQNAILSNKIVIFSKSYCPYCLRSKRIFSQLKEEPFVVE-LDQREDGDQIQY 87
Query 116 YLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIAA 157
L E G R+VP+VF+NG GG DD A +++G LQKL+AA
Sbjct 88 ELLEFVGRRTVPQVFVNGKHIGGSDDLGAALESGQLQKLLAA 129
> YDR513w
Length=143
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 73/139 (52%), Gaps = 14/139 (10%)
Query 20 FINLLYPAAFAGRIWARARLRYNSCRTLFASTMALNSAQDVPQWVDSLVNGHKVTIFAKT 79
I ++ FA RI A+ F ST + S + V V L+ +V + AKT
Sbjct 11 LIVIIIITLFATRIIAKR----------FLSTPKMVSQETVAH-VKDLIGQKEVFVAAKT 59
Query 80 HCPYCQAAIAAL-RSLNV--SDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFINGTFF 136
+CPYC+A ++ L + LNV S V +++ + + IQD L+E +G ++VP V+ING
Sbjct 60 YCPYCKATLSTLFQELNVPKSKALVLELDEMSNGSEIQDALEEISGQKTVPNVYINGKHI 119
Query 137 GGGDDTVAGVKNGTLQKLI 155
GG D KNG L +++
Sbjct 120 GGNSDLETLKKNGKLAEIL 138
> CE04353
Length=149
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 34/114 (29%), Positives = 63/114 (55%), Gaps = 3/114 (2%)
Query 46 TLFASTMALNSAQDVPQWVDSLVNG---HKVTIFAKTHCPYCQAAIAALRSLNVSDMHVE 102
T+ A + + D +VN HKV +++KT+CP+ + A L + + DM +
Sbjct 19 TVHAELSKKKEDKTLKDLEDKIVNDVMTHKVMVYSKTYCPWSKRLKAILANYEIDDMKIV 78
Query 103 QIENNPHCNAIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
+++ + +Q+ LK+ +G +VP++FI+G F GG D+T A + G L+ L+
Sbjct 79 ELDRSNQTEEMQEILKKYSGRTTVPQLFISGKFVGGHDETKAIEEKGELRPLLE 132
> Hs4504025
Length=106
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 51/92 (55%), Gaps = 2/92 (2%)
Query 62 QWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVE--QIENNPHCNAIQDYLKE 119
++V+ + KV +F K CPYC+ A L L + +E I H N IQDYL++
Sbjct 4 EFVNCKIQPGKVVVFIKPTCPYCRRAQEILSQLPIKQGLLEFVDITATNHTNEIQDYLQQ 63
Query 120 KTGARSVPRVFINGTFFGGGDDTVAGVKNGTL 151
TGAR+VPRVFI GG D V+ ++G L
Sbjct 64 LTGARTVPRVFIGKDCIGGCSDLVSLQQSGEL 95
> SPAC4F10.20
Length=101
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 37/95 (38%), Positives = 51/95 (53%), Gaps = 1/95 (1%)
Query 60 VPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKE 119
V +VDS V + V +FAK++CPYC A + + V QI+ + + IQ YL +
Sbjct 4 VESFVDSAVADNDVVVFAKSYCPYCHATEKVIADKKIK-AQVYQIDLMNNGDEIQSYLLK 62
Query 120 KTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKL 154
KTG R+VP +FI+ GG D A K G L L
Sbjct 63 KTGQRTVPNIFIHQKHVGGNSDFQALFKKGELDSL 97
> Hs21361507
Length=165
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 51/92 (55%), Gaps = 1/92 (1%)
Query 64 VDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGA 123
+ ++ + V IF+KT C YC A +NV + V +++ + N QD L + TG
Sbjct 61 IQETISDNCVVIFSKTSCSYCTMAKKLFHDMNV-NYKVVELDLLEYGNQFQDALYKMTGE 119
Query 124 RSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
R+VPR+F+NGTF GG DT K G L L+
Sbjct 120 RTVPRIFVNGTFIGGATDTHRLHKEGKLLPLV 151
> HsM7705628
Length=151
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 51/92 (55%), Gaps = 1/92 (1%)
Query 64 VDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGA 123
+ ++ + V IF+KT C YC A +NV + V +++ + N QD L + TG
Sbjct 47 IQETISDNCVVIFSKTSCSYCTMAKKLFHDMNV-NYKVVELDLLEYGNQFQDALYKMTGE 105
Query 124 RSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
R+VPR+F+NGTF GG DT K G L L+
Sbjct 106 RTVPRIFVNGTFIGGATDTHRLHKEGKLLPLV 137
> Hs22048950
Length=106
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 50/92 (54%), Gaps = 2/92 (2%)
Query 62 QWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVE--QIENNPHCNAIQDYLKE 119
++V+ + K+ +F K CPYC+ A L L+ +E I + H N IQDYL++
Sbjct 4 EFVNCKIQPGKLVVFIKPTCPYCRRAQEILSQLSTKQRLLEFVDITASNHTNKIQDYLQQ 63
Query 120 KTGARSVPRVFINGTFFGGGDDTVAGVKNGTL 151
TGAR VPRVFI GG D V+ + G L
Sbjct 64 LTGARMVPRVFIGKDCIGGCSDLVSMQQIGEL 95
> SPAC15E1.09
Length=110
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 52/96 (54%), Gaps = 5/96 (5%)
Query 63 WVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNV--SDMHVEQIENNPHCNAIQDYLKEK 120
+V+ ++ + VT+F+K+ CP+C+AA L + +++IEN IQ YL EK
Sbjct 8 FVEKAISNNPVTVFSKSFCPFCKAAKNTLTKYSAPYKAYELDKIENGSD---IQAYLHEK 64
Query 121 TGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
T +VP +F F GG D +GTL K+IA
Sbjct 65 TKQSTVPSIFFRNQFIGGNSDLNKLRSSGTLTKMIA 100
> At5g40370
Length=111
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 47/94 (50%), Gaps = 1/94 (1%)
Query 62 QWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKT 121
Q +VN V +F+KT+CPYC L+ L VE ++ + IQ L E T
Sbjct 4 QKAKEIVNSESVVVFSKTYCPYCVRVKELLQQLGAKFKAVE-LDTESDGSQIQSGLAEWT 62
Query 122 GARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
G R+VP VFI G GG D T K+G L L+
Sbjct 63 GQRTVPNVFIGGNHIGGCDATSNLHKDGKLVPLL 96
> YCL035c
Length=110
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 32/88 (36%), Positives = 49/88 (55%), Gaps = 3/88 (3%)
Query 57 AQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAAL-RSLNV--SDMHVEQIENNPHCNAI 113
+Q+ + V L+ +++ + +KT+CPYC AA+ L L V S + V Q+ + I
Sbjct 3 SQETIKHVKDLIAENEIFVASKTYCPYCHAALNTLFEKLKVPRSKVLVLQLNDMKEGADI 62
Query 114 QDYLKEKTGARSVPRVFINGTFFGGGDD 141
Q L E G R+VP ++ING GG DD
Sbjct 63 QAALYEINGQRTVPNIYINGKHIGGNDD 90
> At5g63030
Length=111
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 44/90 (48%), Gaps = 1/90 (1%)
Query 66 SLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARS 125
+V+ + V +F+KT+C YCQ L L + V +++ IQ L E TG +
Sbjct 10 EIVSAYPVVVFSKTYCGYCQRVKQLLTQLGAT-FKVLELDEMSDGGEIQSALSEWTGQTT 68
Query 126 VPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
VP VFI G GG D + K G L L+
Sbjct 69 VPNVFIKGNHIGGCDRVMETNKQGKLVPLL 98
> At2g20270
Length=179
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 48/93 (51%), Gaps = 0/93 (0%)
Query 64 VDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGA 123
V + V + V +++KT C Y + +SL V + VE + + +Q+ L++ TG
Sbjct 78 VKTTVAENPVVVYSKTWCSYSSQVKSLFKSLQVEPLVVELDQLGSEGSQLQNVLEKITGQ 137
Query 124 RSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
+VP VFI G GG DT+ G L+ ++A
Sbjct 138 YTVPNVFIGGKHIGGCSDTLQLHNKGELEAILA 170
> At3g54900
Length=173
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 51/104 (49%), Gaps = 12/104 (11%)
Query 60 VPQWVDSL---VNGHKVTIFAK-----THCPYCQAAIAALRSLNVSDMHVEQIENNPHCN 111
PQ D+L VN KV +F K C + + L++LNV V +EN
Sbjct 68 TPQLKDTLEKLVNSEKVVLFMKGTRDFPMCGFSNTVVQILKNLNVPFEDVNILENE---- 123
Query 112 AIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
++ LKE + + P+++I G FFGG D T+ K G LQ+ +
Sbjct 124 MLRQGLKEYSNWPTFPQLYIGGEFFGGCDITLEAFKTGELQEEV 167
> At3g62960
Length=102
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 42/83 (50%), Gaps = 1/83 (1%)
Query 73 VTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFIN 132
V IF K+ C C A R L V + +I+N+P C I+ L A +VP VF++
Sbjct 13 VVIFTKSSCCLCYAVQILFRDLRVQPT-IHEIDNDPDCREIEKALVRLGCANAVPAVFVS 71
Query 133 GTFFGGGDDTVAGVKNGTLQKLI 155
G G +D ++ +G+L LI
Sbjct 72 GKLVGSTNDVMSLHLSGSLVPLI 94
> At2g47880
Length=102
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 41/83 (49%), Gaps = 1/83 (1%)
Query 73 VTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFIN 132
V IF K+ C C A R L V + +I+N+P C I+ L + +VP VF+
Sbjct 13 VVIFTKSSCCLCYAVQILFRDLRVQPT-IHEIDNDPDCREIEKALLRLGCSTAVPAVFVG 71
Query 133 GTFFGGGDDTVAGVKNGTLQKLI 155
G G ++ ++ +G+L LI
Sbjct 72 GKLVGSTNEVMSLHLSGSLVPLI 94
> YBR014c
Length=203
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 42/79 (53%), Gaps = 2/79 (2%)
Query 75 IFAKTHCPYCQAAIAAL-RSLNVS-DMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFIN 132
+F+KT CPY + A L S S +V +++ + H +QD +++ TG R+VP V I
Sbjct 102 VFSKTGCPYSKKLKALLTNSYTFSPSYYVVELDRHEHTKELQDQIEKVTGRRTVPNVIIG 161
Query 133 GTFFGGGDDTVAGVKNGTL 151
GT GG + KN L
Sbjct 162 GTSRGGYTEIAELHKNDEL 180
> At2g38270
Length=293
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 45/99 (45%), Gaps = 6/99 (6%)
Query 62 QWVDSLVNGHKVTIFAK-----THCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDY 116
+ +D LV KV F K C + Q + L S V D + ++ + + +++
Sbjct 195 ELIDRLVKESKVVAFIKGSRSAPQCGFSQRVVGILESQGV-DYETVDVLDDEYNHGLRET 253
Query 117 LKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
LK + + P++F+ G GG D + +NG L ++
Sbjct 254 LKNYSNWPTFPQIFVKGELVGGCDILTSMYENGELANIL 292
> 7293005_2
Length=116
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 47/94 (50%), Gaps = 11/94 (11%)
Query 64 VDSLVNGHKVTIFAKTH-----CPYCQAAIAALRSLNVS-DMHVEQIENNPHCNAIQDYL 117
+D LV +KV +F K + C + A + +R V D H + ++N ++DY
Sbjct 1 MDKLVRTNKVVVFMKGNPQAPRCGFSNAVVQIMRMHGVQYDAH-DVLQNESLRQGVKDY- 58
Query 118 KEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTL 151
T ++P+VFING F GG D + ++G L
Sbjct 59 ---TDWPTIPQVFINGEFVGGCDILLQMHQSGDL 89
> YDL010w
Length=231
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/71 (25%), Positives = 39/71 (54%), Gaps = 2/71 (2%)
Query 73 VTIFAKTHCPYCQAAIAALRSLN--VSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVF 130
+ IF+K+ C Y + L + + + ++ +++ + H +Q+Y+K TG +VP +
Sbjct 128 IIIFSKSTCSYSKGMKELLENEYQFIPNYYIIELDKHGHGEELQEYIKLVTGRGTVPNLL 187
Query 131 INGTFFGGGDD 141
+NG GG ++
Sbjct 188 VNGVSRGGNEE 198
> SPCC1450.06c
Length=166
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 38/85 (44%), Gaps = 2/85 (2%)
Query 73 VTIFAKTHCPYCQAAIAALRSLNVSDMH--VEQIENNPHCNAIQDYLKEKTGARSVPRVF 130
V IF++ CPY AA L D V ++ + H ++D+L + ++P +F
Sbjct 68 VIIFSRPGCPYSAAAKKLLTETLRLDPPAVVVEVTDYEHTQELRDWLSSISDISTMPNIF 127
Query 131 INGTFFGGGDDTVAGVKNGTLQKLI 155
+ G GG D A + LQ +
Sbjct 128 VGGHSIGGSDSVRALYQEEKLQSTL 152
> At4g28730
Length=176
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 43/91 (47%), Gaps = 2/91 (2%)
Query 68 VNGHKVTIFAKTHCPYCQAAIAALRS--LNVSDMHVEQIENNPHCNAIQDYLKEKTGARS 125
V + V I++KT + + R N+ + VE + P +Q L+ TG +
Sbjct 77 VTENTVVIYSKTWICFISFFLLNWRMGLSNILPLVVELDQLGPQGPQLQKVLERLTGQHT 136
Query 126 VPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
VP VF+ G GG DTV + G L+ ++A
Sbjct 137 VPNVFVCGKHIGGCTDTVKLNRKGDLELMLA 167
> At1g06830
Length=99
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 39/83 (46%), Gaps = 1/83 (1%)
Query 73 VTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFIN 132
V IF K+ C A + L V+ + +I+ +P C I+ L ++ VP VFI
Sbjct 13 VVIFTKSSCCLSYAVQVLFQDLGVNP-KIHEIDKDPECREIEKALMRLGCSKPVPAVFIG 71
Query 133 GTFFGGGDDTVAGVKNGTLQKLI 155
G G ++ ++ + +L L+
Sbjct 72 GKLVGSTNEVMSMHLSSSLVPLV 94
> At2g30540
Length=102
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 39/83 (46%), Gaps = 1/83 (1%)
Query 73 VTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFIN 132
V IF+K+ C A + L V V +I+ +P C I+ L + VP +F+
Sbjct 13 VVIFSKSSCCMSYAVQVLFQDLGVHPT-VHEIDKDPECREIEKALMRLGCSTPVPAIFVG 71
Query 133 GTFFGGGDDTVAGVKNGTLQKLI 155
G G ++ ++ +G+L L+
Sbjct 72 GKLIGSTNEVMSLHLSGSLVPLV 94
> At4g04950
Length=488
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 25/119 (21%), Positives = 57/119 (47%), Gaps = 9/119 (7%)
Query 39 LRYNSCRTLFASTMALNSAQDVPQWVDSLVNGHKVTIFAK-----THCPYCQAAIAALRS 93
++ N+ +L +++A + ++ L N H V +F K C + + + L+
Sbjct 132 VKENAKASLQDRAQPVSTADALKSRLEKLTNSHPVMLFMKGIPEEPRCGFSRKVVDILKE 191
Query 94 LNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQ 152
+NV + + +N +++ LK+ + + P+++ NG GG D +A ++G L+
Sbjct 192 VNVDFGSFDILSDN----EVREGLKKFSNWPTFPQLYCNGELLGGADIAIAMHESGELK 246
Score = 28.1 bits (61), Expect = 7.9, Method: Composition-based stats.
Identities = 25/98 (25%), Positives = 44/98 (44%), Gaps = 11/98 (11%)
Query 64 VDSLVNGHKVTIFAKTHC--PYCQAAIAALRSLNVSDMHVEQIENNPHC----NAIQDYL 117
++ LVN V +F K P C + + LN E+IE + ++ L
Sbjct 287 LEGLVNSKPVMLFMKGRPEEPKCGFSGKVVEILNQ-----EKIEFGSFDILLDDEVRQGL 341
Query 118 KEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
K + S P++++ G GG D + K+G L+K++
Sbjct 342 KVYSNWSSYPQLYVKGELMGGSDIVLEMQKSGELKKVL 379
> SPBC26H8.06
Length=244
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 24/102 (23%), Positives = 49/102 (48%), Gaps = 9/102 (8%)
Query 59 DVPQWVDSLVNGHKVTIFAKTH-----CPYCQAAIAALRSLNVSDMHVEQIENNPHCNAI 113
++ + + +L N H V +F K C + + + LR NV + ++ ++
Sbjct 145 ELNERLSTLTNAHNVMLFLKGTPSEPACGFSRKLVGLLREQNVQYGFFNILADD----SV 200
Query 114 QDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
+ LK + + P+++I G F GG D ++NG LQ+++
Sbjct 201 RQGLKVFSDWPTFPQLYIKGEFVGGLDIVSEMIENGELQEML 242
> SPAPB2B4.02
Length=146
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 23/98 (23%), Positives = 53/98 (54%), Gaps = 4/98 (4%)
Query 62 QWVDSLVNGHKVTIFAK--THCPYCQAAIAALRSLNVSDMHVEQI--ENNPHCNAIQDYL 117
Q ++ V + +F K P C ++ A++ L++ ++ +++ N + +++ +
Sbjct 27 QALEQAVKEDPIVLFMKGTPTRPMCGFSLKAIQILSLENVASDKLVTYNVLSNDELREGI 86
Query 118 KEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
KE + ++P+++ING F GG D + K+G L K++
Sbjct 87 KEFSDWPTIPQLYINGEFVGGSDILASMHKSGELHKIL 124
> At2g47870
Length=103
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 22/97 (22%), Positives = 45/97 (46%), Gaps = 2/97 (2%)
Query 62 QWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKT 121
+ V L + IF K+ C C + L S + +++ +P ++ L
Sbjct 2 ERVRDLASEKAAVIFTKSSCCMCHSIKTLFYELGASPA-IHELDKDPQGPDMERALFRVF 60
Query 122 GAR-SVPRVFINGTFFGGGDDTVAGVKNGTLQKLIAA 157
G+ +VP VF+ G + G D ++ +G+L++++ A
Sbjct 61 GSNPAVPAVFVGGRYVGSAKDVISFHVDGSLKQMLKA 97
> At5g06470
Length=239
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 122 GARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIAA 157
G + PR+FI G + GG ++ VA +NG L+KL+
Sbjct 143 GKVTSPRLFIRGRYIGGAEEVVALNENGKLKKLLQG 178
> ECU09g1375
Length=98
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 32/72 (44%), Gaps = 4/72 (5%)
Query 77 AKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFINGTFF 136
+K CP+C+ A L+S + ++ EN I+ + G + P++F+ F
Sbjct 19 SKDGCPFCKDASEILKSWKIEFACIKNTENERLSKEIE----REYGFSTFPKIFLKRKFV 74
Query 137 GGGDDTVAGVKN 148
GG D VK
Sbjct 75 GGASDLKEYVKK 86
> At3g62950
Length=103
Score = 35.8 bits (81), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 19/95 (20%), Positives = 44/95 (46%), Gaps = 2/95 (2%)
Query 62 QWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKT 121
+ + L + IF K+ C C + L S + +++ +P ++ L+
Sbjct 2 ERIRDLSSKKAAVIFTKSSCCMCHSIKTLFYELGASPA-IHELDKDPEGREMERALRALG 60
Query 122 GAR-SVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
+ +VP VF+ G + G D ++ +G+L++++
Sbjct 61 SSNPAVPAVFVGGRYIGSAKDIISFHVDGSLKQML 95
> At3g62930
Length=102
Score = 35.0 bits (79), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 29/87 (33%), Positives = 42/87 (48%), Gaps = 4/87 (4%)
Query 64 VDSLVNGHKVTIFAKTHCPYCQA-AIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTG 122
V SLV V IF+K+ C C + +I L S + M V +++ + I+ L +
Sbjct 4 VRSLVEDKPVVIFSKSSC--CMSHSIQTLISGFGAKMTVYELDQFSNGQEIEKALVQMGC 61
Query 123 ARSVPRVFINGTFFGGGDDTVA-GVKN 148
SVP VFI F GG + + VKN
Sbjct 62 KPSVPAVFIGQQFIGGANQVMTLQVKN 88
> At3g02000
Length=136
Score = 34.7 bits (78), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 45/98 (45%), Gaps = 7/98 (7%)
Query 64 VDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYL------ 117
++SL + V IF+ + C C A R + VS V +++ +P+ IQ L
Sbjct 32 IESLASESAVVIFSVSTCCMCHAVKGLFRGMGVSPA-VHELDLHPYGGDIQRALIRLLGC 90
Query 118 KEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
+ S+P VFI G G D +A NG+L L+
Sbjct 91 SGSSSPGSLPVVFIGGKLVGAMDRVMASHINGSLVPLL 128
> Hs6912400
Length=822
Score = 34.3 bits (77), Expect = 0.098, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 29/64 (45%), Gaps = 15/64 (23%)
Query 31 GRIWARARLRYNSC------RTLFASTMALNSAQDVPQWVDSLVNGHKVTIFAKTHCPYC 84
G IW R L Y SC R L ++A + A + P W+ L+ ++ CP+C
Sbjct 767 GHIWLRCFLTYQSCQSLIYRRCLLHDSIARHPAPEDPDWIKRLL---------QSPCPFC 817
Query 85 QAAI 88
+ +
Sbjct 818 DSPV 821
> YLR364w
Length=109
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 43/93 (46%), Gaps = 5/93 (5%)
Query 65 DSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSD-MHVEQIENNPHCNAIQDY---LKEK 120
+ ++ H + + CP C A + LNV D + V I + P N + + ++
Sbjct 9 EEMIKSHPYFQLSASWCPDCVYANSIWNKLNVQDKVFVFDIGSLPR-NEQEKWRIAFQKV 67
Query 121 TGARSVPRVFINGTFFGGGDDTVAGVKNGTLQK 153
G+R++P + +NG F+G GTL++
Sbjct 68 VGSRNLPTIVVNGKFWGTESQLHRFEAKGTLEE 100
> At3g15660
Length=169
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 45/104 (43%), Gaps = 9/104 (8%)
Query 55 NSAQDVPQWVDSLVNGHKVTIFAK-----THCPYCQAAIAALRSLNVSDMHVEQIENNPH 109
+S + V++ V + V I+ K C + A+ L+ NV +E+
Sbjct 60 DSTDSLKDIVENDVKDNPVMIYMKGVPESPQCGFSSLAVRVLQQYNVPISSRNILEDQEL 119
Query 110 CNAIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQK 153
NA++ + T P++FI G F GG D + K G L++
Sbjct 120 KNAVKSFSHWPT----FPQIFIKGEFIGGSDIILNMHKEGELEQ 159
> At4g33040
Length=144
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 52/109 (47%), Gaps = 9/109 (8%)
Query 49 ASTMALNSAQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNV--SDMHVEQIEN 106
+++++++ + + L++ H V IF+++ C C L ++ V + + ++ E
Sbjct 27 STSLSIDEEESTEAKIRRLISEHPVIIFSRSSCCMCHVMKRLLATIGVIPTVIELDDHEV 86
Query 107 NPHCNAIQDYLKEKTGARSV----PRVFINGTFFGGGDDTVAGVKNGTL 151
+ A+QD E +G SV P VFI GG + VA +G L
Sbjct 87 SSLPTALQD---EYSGGVSVVGPPPAVFIGRECVGGLESLVALHLSGQL 132
> At4g10630
Length=334
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 20/78 (25%), Positives = 36/78 (46%), Gaps = 4/78 (5%)
Query 84 CQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSV----PRVFINGTFFGGG 139
C A AA+ S V + + + + ++ G V PRVF+ G + GGG
Sbjct 195 CNAVRAAVESFGVVVCERDVSMDRRFREELVSLMAKRVGDEGVAALPPRVFVKGRYIGGG 254
Query 140 DDTVAGVKNGTLQKLIAA 157
++ + V+ G+ +LI+
Sbjct 255 EEVLRLVEEGSFGELISG 272
> At5g14070
Length=140
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 43/100 (43%), Gaps = 9/100 (9%)
Query 64 VDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGA 123
++S+ + V IF+ + C C A R + VS V +++ P+ I L G
Sbjct 34 IESMAAENAVVIFSVSTCCMCHAIKRLFRGMGVSPA-VHELDLLPYGVEIHRALLRLLGC 92
Query 124 RS--------VPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
S +P VFI G G + +A NG+L L+
Sbjct 93 SSGGATSPGALPVVFIGGKMVGAMERVMASHINGSLVPLL 132
> At1g03850
Length=90
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 41/81 (50%), Gaps = 9/81 (11%)
Query 73 VTIFAKTHCPYCQAAIAALRSLNVSDMHVE--QIENNPHCNAIQDYLKEKTGARSVPRVF 130
V +FA+ C A L + V+ + VE + +NN + N + D KEK +P ++
Sbjct 5 VVVFARRGCCLGHVAKRLLLTHGVNPVVVEIGEEDNNNYDNIVSD--KEK-----LPMMY 57
Query 131 INGTFFGGGDDTVAGVKNGTL 151
I G FGG ++ +A NG L
Sbjct 58 IGGKLFGGLENLMAAHINGDL 78
> At3g11920
Length=630
Score = 32.7 bits (73), Expect = 0.31, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 36/69 (52%), Gaps = 8/69 (11%)
Query 72 KVTIFAKTHCPYCQAAIAALRS--LNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRV 129
++T F++++C A LR + S+++++ + + L E+TG+ VP++
Sbjct 158 RITFFSRSNCRDSTAVRLFLRERGFDFSEINIDVYSSR------EKELVERTGSSQVPQI 211
Query 130 FINGTFFGG 138
F N FGG
Sbjct 212 FFNEKHFGG 220
> YPL059w
Length=150
Score = 32.0 bits (71), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 26/135 (19%), Positives = 63/135 (46%), Gaps = 19/135 (14%)
Query 28 AFAGRIWARARLRYNSCRTLFASTMALNSAQDVPQWVDSLVNGHKVTIFAK-----THCP 82
+F+ + A+ LRY + ++ ST ++ + ++ + V +F K C
Sbjct 11 SFSPILRAKTLLRYQN--RMYLST-------EIRKAIEDAIESAPVVLFMKGTPEFPKCG 61
Query 83 YCQAAIAAL--RSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFINGTFFGGGD 140
+ +A I L + ++ + + +P +++ +KE + ++P++++N F GG D
Sbjct 62 FSRATIGLLGNQGVDPAKFAAYNVLEDPE---LREGIKEFSEWPTIPQLYVNKEFIGGCD 118
Query 141 DTVAGVKNGTLQKLI 155
+ ++G L L+
Sbjct 119 VITSMARSGELADLL 133
> At1g32760
Length=314
Score = 31.6 bits (70), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 119 EKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
E T A PRVF+ G + GG ++ + V+ G L +L+
Sbjct 216 ESTAAVLPPRVFVKGKYIGGAEEVMRLVEEGLLGELL 252
> At3g28850
Length=428
Score = 31.6 bits (70), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 44/87 (50%), Gaps = 12/87 (13%)
Query 78 KTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGAR-------SVPRVF 130
KT+ C + L+SL + V++ + + H + +D LKE G + ++PRVF
Sbjct 265 KTYEESCDVRVI-LKSLGI---RVDERDVSMH-SGFKDELKELLGEKFNKGVGITLPRVF 319
Query 131 INGTFFGGGDDTVAGVKNGTLQKLIAA 157
+ + GG ++ ++G L+KL+
Sbjct 320 LGRKYIGGAEEIRKLNEDGKLEKLLGG 346
> CE29627
Length=119
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 43/100 (43%), Gaps = 10/100 (10%)
Query 64 VDSLVNGHKVTIFAKTHCPYCQAAI-------AALRSLNVSDMHVEQIENNPHCNAIQDY 116
V V V ++ KT C +C A A + +N+ + Q ++ I +
Sbjct 13 VQEQVKKDPVVMYTKTSCTFCNRAKDLFSDVRVAYKEVNLDTLKASQPDDYL---GIVNG 69
Query 117 LKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
L T SVP++F+ G F GG + A +G L + IA
Sbjct 70 LVYTTRQTSVPQIFVCGRFIGGYTELDALRNSGHLFEAIA 109
> At1g75050
Length=257
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 17 PGAFINLLYPAAFAGRIWARARLRYNS 43
PGA + L PA ++GR WAR +++
Sbjct 69 PGASVQLTAPAGWSGRFWARTGCNFDA 95
Lambda K H
0.324 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2106641548
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40