bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2495_orf3
Length=238
Score E
Sequences producing significant alignments: (Bits) Value
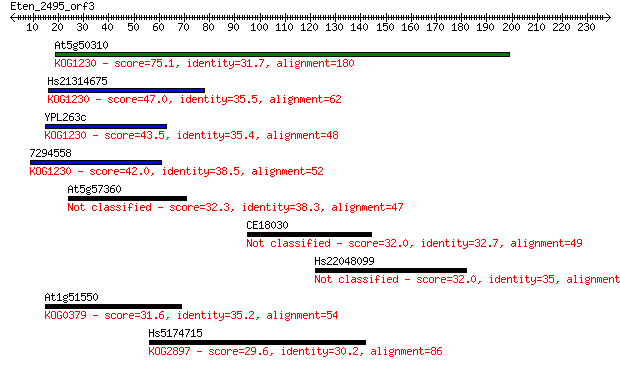
At5g50310 75.1 1e-13
Hs21314675 47.0 3e-05
YPL263c 43.5 3e-04
7294558 42.0 0.001
At5g57360 32.3 0.95
CE18030 32.0 1.2
Hs22048099 32.0 1.3
At1g51550 31.6 1.5
Hs5174715 29.6 6.2
> At5g50310
Length=596
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 57/190 (30%), Positives = 92/190 (48%), Gaps = 14/190 (7%)
Query 19 PLPRLHGMLCVRGSNLVLMGGIMELGSKEITLDDCWTLNLNKRDRWIRVLEGTMHEQEW- 77
P R++ + V L + GG+ME+ KE+TLDD ++LNL+K D W ++ T E EW
Sbjct 369 PCGRINSCMVVGKDTLYIYGGMMEIKDKEVTLDDLYSLNLSKLDEWKCIIPTT--ETEWV 426
Query 78 -------QGADSDHESSNESDGDVSSSSGSSSLESDELD-DGTSESDEEVPRRQGNVQRV 129
D D + S + S E + +D DG+ + V +G + +
Sbjct 427 EVSDDEEGDEDDDEDDSEDEGNSEESDDEDDDEEVEAMDVDGSVKVGVVVAMIKGQGKSL 486
Query 130 RRRVQ-EEIQQLREQYDLDDPLETPAQGECLRDFFDRTRAHWIEKVSTAGNVVRNSKELT 188
RR+ + I+Q+R L D TP GE L+DF+ RT +W +++ + KEL
Sbjct 487 RRKEKRARIEQIRANLGLSDSQRTPVPGETLKDFYKRTNMYW--QMAAYEHTQHTGKELR 544
Query 189 REAFEAASAR 198
++ F+ A R
Sbjct 545 KDGFDLAETR 554
> Hs21314675
Length=520
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 35/62 (56%), Gaps = 0/62 (0%)
Query 16 ADMPLPRLHGMLCVRGSNLVLMGGIMELGSKEITLDDCWTLNLNKRDRWIRVLEGTMHEQ 75
A P PR + ML V+ L + GG+ E G +++TL D L+L++ + W ++E Q
Sbjct 426 APGPCPRSNAMLAVKHGVLYVYGGMFEAGDRQVTLSDLHCLDLHRMEAWKALVEMDPETQ 485
Query 76 EW 77
EW
Sbjct 486 EW 487
> YPL263c
Length=651
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 29/48 (60%), Gaps = 0/48 (0%)
Query 15 SADMPLPRLHGMLCVRGSNLVLMGGIMELGSKEITLDDCWTLNLNKRD 62
S +P PR + CV G +L + G+ ELG K+ ++ ++++LNK D
Sbjct 447 SNQLPHPRFNAATCVVGDSLFIYSGVWELGEKDYPINSFYSIDLNKLD 494
> 7294558
Length=509
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 33/56 (58%), Gaps = 4/56 (7%)
Query 9 KTPPLFS----ADMPLPRLHGMLCVRGSNLVLMGGIMELGSKEITLDDCWTLNLNK 60
K P LF+ ++P PR++ +CV L + GGI E K++T +D + L+L+K
Sbjct 409 KIPSLFAKPKPTNVPSPRMNPGMCVCKGTLYIFGGIFEEDDKQLTYNDFYALDLHK 464
> At5g57360
Length=609
Score = 32.3 bits (72), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 4/47 (8%)
Query 24 HGMLCVRGSNLVLMGGIMELGSKEITLDDCWTLNLNKRDRWIRVLEG 70
H + CV GSNLV+ GG + G L+D + LNL+ + R + G
Sbjct 348 HTLTCVNGSNLVVFGGCGQQG----LLNDVFVLNLDAKPPTWREISG 390
> CE18030
Length=798
Score = 32.0 bits (71), Expect = 1.2, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 95 SSSSGSSSLESDELDDGTSESDEEVPRRQGNVQRVRRRVQEEIQQLREQ 143
+ SSG S E D+G E +EE P+++ V++V + +QQ + Q
Sbjct 740 ADSSGDDSEEVASSDEGIQEVEEEQPKQKKLVKKVEPKTIPRVQQKKPQ 788
> Hs22048099
Length=442
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 34/60 (56%), Gaps = 4/60 (6%)
Query 122 RQGNVQRVRRRVQEEIQQLREQYDLDDPLETPAQGECLRDFFDRTRAHWIEKVSTAGNVV 181
R+ VQ+ ++ ++EEIQ LR Y L L +Q E L+D F+ T + + E + N+V
Sbjct 322 RETAVQQYKK-LEEEIQTLRVYYSLHKSL---SQEENLKDQFNYTLSTYEEALKNRENIV 377
> At1g51550
Length=478
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 31/57 (54%), Gaps = 6/57 (10%)
Query 15 SADMPLPRL-HGMLCVRGSNLVLMGGIMELGSKEITLDDCWTLNLNK--RDRWIRVL 68
S +P PR H + C+R + +VL GG G LDD W L++ + ++WI++
Sbjct 282 SPQLPPPRSGHTLTCIRENQVVLFGG---RGLGYDVLDDVWILDIQEPCEEKWIQIF 335
> Hs5174715
Length=364
Score = 29.6 bits (65), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 42/90 (46%), Gaps = 17/90 (18%)
Query 56 LNLNKRDRWIRVLEGTMHEQ----EWQGADSDHESSNESDGDVSSSSGSSSLESDELDDG 111
L + D + + G E+ E+QG SD E +SD D+ DE D+
Sbjct 21 LEAEEEDEFYQTTYGGFTEESGDDEYQGDQSDTEDEVDSDFDI-----------DEGDEP 69
Query 112 TSESDEEVPRRQGNVQRVRRRVQEEIQQLR 141
+S+ + E PRR+ V V + +E ++ LR
Sbjct 70 SSDGEAEEPRRKRRV--VTKAYKEPLKSLR 97
Lambda K H
0.314 0.131 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4740636838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40