bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2481_orf3
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
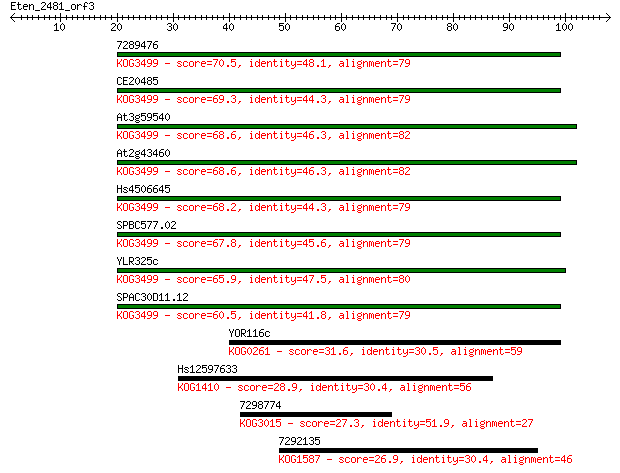
7289476 70.5 8e-13
CE20485 69.3 2e-12
At3g59540 68.6 3e-12
At2g43460 68.6 3e-12
Hs4506645 68.2 4e-12
SPBC577.02 67.8 5e-12
YLR325c 65.9 2e-11
SPAC30D11.12 60.5 8e-10
YOR116c 31.6 0.38
Hs12597633 28.9 2.5
7298774 27.3 6.6
7292135 26.9 8.8
> 7289476
Length=70
Score = 70.5 bits (171), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 38/79 (48%), Positives = 48/79 (60%), Gaps = 15/79 (18%)
Query 20 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 79
M RE++++++FL ARR DAR V I ++ TKFKIRCS+ LYTL
Sbjct 1 MPREIKEVKDFLNKARRSDARAVKIKKN---------------PTNTKFKIRCSRFLYTL 45
Query 80 VVADKAKAQKIEGSLPPSL 98
VV DK KA KI+ SLPP L
Sbjct 46 VVQDKEKADKIKQSLPPGL 64
> CE20485
Length=70
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 51/79 (64%), Gaps = 15/79 (18%)
Query 20 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 79
M +E+++I++FL ARRKDA+ V I ++ TKFK+RC+ +LYTL
Sbjct 1 MPKEIKEIKDFLVKARRKDAKSVKIKKNSNN---------------TKFKVRCASYLYTL 45
Query 80 VVADKAKAQKIEGSLPPSL 98
VVADK KA+K++ SLPP +
Sbjct 46 VVADKDKAEKLKQSLPPGI 64
> At3g59540
Length=69
Score = 68.6 bits (166), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 38/82 (46%), Positives = 50/82 (60%), Gaps = 15/82 (18%)
Query 20 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 79
M +++ +I++FL TARRKDAR V I RS I KFK+RCS++LYTL
Sbjct 1 MPKQIHEIKDFLLTARRKDARSVKIKRS---------------KDIVKFKVRCSRYLYTL 45
Query 80 VVADKAKAQKIEGSLPPSLKQQ 101
V D+ KA K++ SLPP L Q
Sbjct 46 CVFDQEKADKLKQSLPPGLSVQ 67
> At2g43460
Length=69
Score = 68.6 bits (166), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 38/82 (46%), Positives = 50/82 (60%), Gaps = 15/82 (18%)
Query 20 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 79
M +++ +I++FL TARRKDAR V I RS I KFK+RCS++LYTL
Sbjct 1 MPKQIHEIKDFLLTARRKDARSVKIKRS---------------KDIVKFKVRCSRYLYTL 45
Query 80 VVADKAKAQKIEGSLPPSLKQQ 101
V D+ KA K++ SLPP L Q
Sbjct 46 CVFDQEKADKLKQSLPPGLSVQ 67
> Hs4506645
Length=70
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 50/79 (63%), Gaps = 15/79 (18%)
Query 20 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 79
M R++ +I++FL TARRKDA+ V I ++ KFK+RCS++LYTL
Sbjct 1 MPRKIEEIKDFLLTARRKDAKSVKIKKNKDN---------------VKFKVRCSRYLYTL 45
Query 80 VVADKAKAQKIEGSLPPSL 98
V+ DK KA+K++ SLPP L
Sbjct 46 VITDKEKAEKLKQSLPPGL 64
> SPBC577.02
Length=74
Score = 67.8 bits (164), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 36/79 (45%), Positives = 48/79 (60%), Gaps = 14/79 (17%)
Query 20 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 79
M R++ DI++FL+ ARRKDA I ++ + KFK+RCSK+LYTL
Sbjct 1 MPRQISDIKQFLEIARRKDATSARIKKNTNKD--------------VKFKLRCSKYLYTL 46
Query 80 VVADKAKAQKIEGSLPPSL 98
VVAD KA+K+ SLPP L
Sbjct 47 VVADAKKAEKLRQSLPPDL 65
> YLR325c
Length=78
Score = 65.9 bits (159), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 38/82 (46%), Positives = 51/82 (62%), Gaps = 10/82 (12%)
Query 20 MARELRDIREFLQTARRKDARK--VVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLY 77
MARE+ DI++FL+ RR D + V I++ L + G PF TKFK+R S LY
Sbjct 1 MAREITDIKQFLELTRRADVKTATVKINKKLNKA-GKPFRQ-------TKFKVRGSSSLY 52
Query 78 TLVVADKAKAQKIEGSLPPSLK 99
TLV+ D KA+K+ SLPP+LK
Sbjct 53 TLVINDAGKAKKLIQSLPPTLK 74
> SPAC30D11.12
Length=74
Score = 60.5 bits (145), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 33/79 (41%), Positives = 46/79 (58%), Gaps = 14/79 (17%)
Query 20 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 79
M R++ DI+ FLQ A R DA + ++ + KFK+RCS++LYTL
Sbjct 1 MPRQVTDIKLFLQLAHRGDATSARVKKNQNKA--------------VKFKLRCSRYLYTL 46
Query 80 VVADKAKAQKIEGSLPPSL 98
VVAD KA+K+ SLPP+L
Sbjct 47 VVADAKKAEKLRQSLPPAL 65
> YOR116c
Length=1460
Score = 31.6 bits (70), Expect = 0.38, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 36/60 (60%), Gaps = 4/60 (6%)
Query 40 RKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCS-KHLYTLVVADKAKAQKIEGSLPPSL 98
++VV+ + +R KG F+A +AA ++ + ++ S + L+ L +K +A K G+L P +
Sbjct 2 KEVVVSETPKRIKGLEFSALSAADIVAQSEVEVSTRDLFDL---EKDRAPKANGALDPKM 58
> Hs12597633
Length=1088
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 30/62 (48%), Gaps = 6/62 (9%)
Query 31 LQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITK------FKIRCSKHLYTLVVADK 84
L +R D R +++ +PK APF A VI K F+++ + ++ ++AD
Sbjct 77 LPVEQRMDIRNYILNYVASQPKLAPFVIQALIQVIAKITKLGWFEVQKDQFVFREIIADV 136
Query 85 AK 86
K
Sbjct 137 KK 138
> 7298774
Length=279
Score = 27.3 bits (59), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 18/30 (60%), Gaps = 3/30 (10%)
Query 42 VVIHRSLRRPKGAPFAAAA---AAAVITKF 68
V+ HR L+ PKG PF + A AV T+F
Sbjct 56 VLDHRKLKTPKGTPFIVRSEPLAIAVATEF 85
> 7292135
Length=563
Score = 26.9 bits (58), Expect = 8.8, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 25/46 (54%), Gaps = 3/46 (6%)
Query 49 RRPKGAPFAAAAAAAVITKFKIRCSKHLYTLVVADKAKAQKIEGSL 94
R P PF A + A +TK+++ S ++ L ++ + Q+ EG L
Sbjct 414 RNPHITPFLATSQAGAVTKYRLINSPNMLGL---EQQRLQRAEGEL 456
Lambda K H
0.322 0.132 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40