bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2476_orf1
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
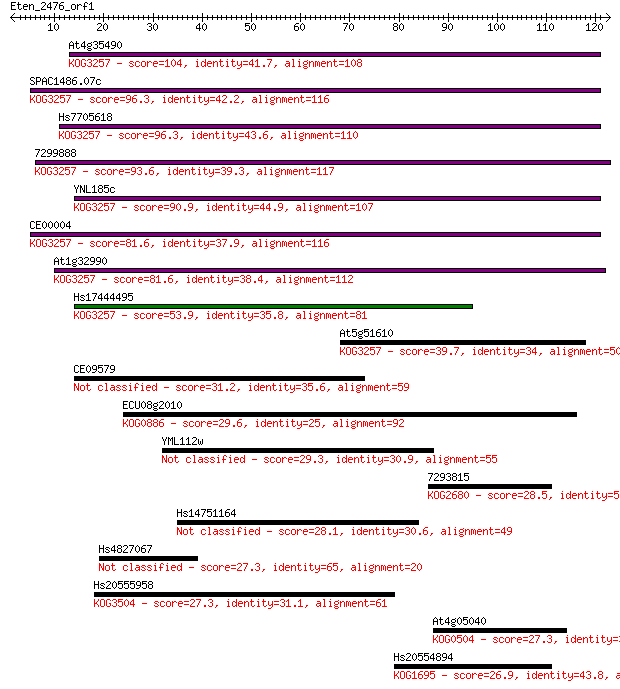
At4g35490 104 4e-23
SPAC1486.07c 96.3 1e-20
Hs7705618 96.3 1e-20
7299888 93.6 8e-20
YNL185c 90.9 6e-19
CE00004 81.6 3e-16
At1g32990 81.6 3e-16
Hs17444495 53.9 6e-08
At5g51610 39.7 0.002
CE09579 31.2 0.55
ECU08g2010 29.6 1.5
YML112w 29.3 2.1
7293815 28.5 3.2
Hs14751164 28.1 4.4
Hs4827067 27.3 7.2
Hs20555958 27.3 7.7
At4g05040 27.3 8.1
Hs20554894 26.9 8.1
> At4g35490
Length=155
Score = 104 bits (260), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 45/108 (41%), Positives = 71/108 (65%), Gaps = 0/108 (0%)
Query 13 FRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRTFK 72
RL VPA A+P+P +G LG +N+MAFCK+FNART K +PD P+ VTI D +F+
Sbjct 17 IRLTVPAGGARPAPPVGPALGQYRLNLMAFCKDFNARTQKYKPDTPMAVTITAFKDNSFE 76
Query 73 FSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMD 120
F++++P+ +W++ +AA GS + G + ++++ +Y IAK K D
Sbjct 77 FTVKSPTVSWYIKKAAGVDKGSTRPGHLSVTTLSVRHVYEIAKVKQTD 124
> SPAC1486.07c
Length=144
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 49/116 (42%), Positives = 64/116 (55%), Gaps = 0/116 (0%)
Query 5 MKMTNLGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIV 64
M T +LIVPA A P+P IG LG G+ + FCKEFNARTA P+ PV I
Sbjct 1 MASTRTTIIKLIVPAGKATPTPPIGPALGARGLKSIDFCKEFNARTAGWMPNTPVPCKIT 60
Query 65 PNADRTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMD 120
RTF F++ TP ++W + + GS ++ G ++LK IY IAK KS D
Sbjct 61 VTPQRTFTFTIHTPPTSWLISKTLDLEKGSSSPLHDIKGQLSLKHIYEIAKLKSTD 116
> Hs7705618
Length=192
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 48/110 (43%), Positives = 65/110 (59%), Gaps = 0/110 (0%)
Query 11 GRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRT 70
G R IV A A P P +G LG G+++ FCKEFN RT I+ +P+ I+ DRT
Sbjct 18 GVIRAIVRAGLAMPGPPLGPVLGQRGVSINQFCKEFNERTKDIKEGIPLPTKILVKPDRT 77
Query 71 FKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMD 120
F+ + P+ ++FL AA G+ Q GKEV G VTLK +Y IA+ K+ D
Sbjct 78 FEIKIGQPTVSYFLKAAAGIEKGARQTGKEVAGLVTLKHVYEIARIKAQD 127
> 7299888
Length=196
Score = 93.6 bits (231), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 46/117 (39%), Positives = 71/117 (60%), Gaps = 0/117 (0%)
Query 6 KMTNLGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVP 65
++T+ + + +PA A P +G LG IN+ AFCK+FNA+TA+++ VP+ I
Sbjct 17 RVTHTSKLKTNIPAGMAAAGPPLGPMLGQRAINIAAFCKDFNAKTAEMKEGVPLPCRISV 76
Query 66 NADRTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMDWP 122
N+DR++ ++ P +T+FL +AA G+ GKEV G +TLK +Y IA K D P
Sbjct 77 NSDRSYDLAIHHPPATFFLKQAAGIQRGTMTPGKEVAGMITLKHLYEIAAIKIQDPP 133
> YNL185c
Length=158
Score = 90.9 bits (224), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 48/120 (40%), Positives = 64/120 (53%), Gaps = 13/120 (10%)
Query 14 RLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRTFKF 73
+LIV A A PSP +G LG GI + FCKEFNAR+A +P VPV V I DRTF F
Sbjct 11 KLIVGAGQAAPSPPVGPALGSKGIKAIDFCKEFNARSANYQPGVPVPVLITIKPDRTFTF 70
Query 74 SLRTPSSTWFLLRAARCPMGSEQAG-------------KEVIGNVTLKEIYHIAKCKSMD 120
+++P + + LL+A + G Q +G ++LK +Y IAK K D
Sbjct 71 EMKSPPTGYLLLKALKMDKGHGQPNVGTMLGSAPAKGPTRALGELSLKHVYEIAKIKKSD 130
> CE00004
Length=195
Score = 81.6 bits (200), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 44/117 (37%), Positives = 66/117 (56%), Gaps = 3/117 (2%)
Query 5 MKMTNLGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIV 64
+K+ + R + A A +P +G LG G+N+ FCKEFN T + VP+ I
Sbjct 15 VKVVHGALLRTNIKAQMASAAPPLGPQLGQRGLNVANFCKEFNKETGHFKQGVPLPTRIT 74
Query 65 PNADRTFKFSLRTPSSTWFLLRAARCPMGSEQAGK-EVIGNVTLKEIYHIAKCKSMD 120
DRT+ + TP++TW L +AA +G +A K E++G +T+K +Y IAK KS D
Sbjct 75 VKPDRTYDLEICTPTTTWLLKQAA--GIGRGKASKDEIVGKLTVKHLYEIAKIKSRD 129
> At1g32990
Length=222
Score = 81.6 bits (200), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 43/113 (38%), Positives = 70/113 (61%), Gaps = 3/113 (2%)
Query 10 LGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTA-KIRPDVPVQVTIVPNAD 68
+G +L + A A P+P +G LG G+N+MAFCK++NARTA K +PV++T+ D
Sbjct 75 VGVIKLALEAGKATPAPPVGPALGSKGVNIMAFCKDYNARTADKAGYIIPVEITVF--DD 132
Query 69 RTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMDW 121
++F F L+TP ++ LL+AA GS+ ++ +G +T+ ++ IA K D
Sbjct 133 KSFTFILKTPPASVLLLKAAGVEKGSKDPQQDKVGVITIDQLRTIAAEKLPDL 185
> Hs17444495
Length=130
Score = 53.9 bits (128), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 42/81 (51%), Gaps = 0/81 (0%)
Query 14 RLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRTFKF 73
R IV A A P P +G LG ++ FCKEFN RT I+ +P+ I D TF+
Sbjct 21 RTIVRAGLAMPGPPLGPVLGQRRASINQFCKEFNERTKDIKEGIPLLTKIFLKPDGTFEI 80
Query 74 SLRTPSSTWFLLRAARCPMGS 94
+ P+ ++F+ AA G+
Sbjct 81 KIGQPTVSYFVKAAAGIEKGA 101
> At5g51610
Length=182
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 32/50 (64%), Gaps = 0/50 (0%)
Query 68 DRTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCK 117
D++F F L+TP +++ LL+AA GS+ + +G +T+ ++ IA+ K
Sbjct 92 DQSFTFILKTPPTSFLLLKAAGVKKGSKDPKQIKVGMITIDQVRKIAEVK 141
> CE09579
Length=1307
Score = 31.2 bits (69), Expect = 0.55, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 14/64 (21%)
Query 14 RLIVPAANAKPSPSIGQTLGP-----LGINMMAFCKEFNARTAKIRPDVPVQVTIVPNAD 68
RL+V K +PS+ TLGP L I++++ CK+F I PNA
Sbjct 1152 RLLVMGKYIKKNPSLALTLGPEDAAKLNIDVVSLCKKFEILYLSI---------AAPNAS 1202
Query 69 RTFK 72
+FK
Sbjct 1203 PSFK 1206
> ECU08g2010
Length=166
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 23/95 (24%), Positives = 46/95 (48%), Gaps = 7/95 (7%)
Query 24 PSPSIGQTLGPLGINMMAFCKEFNARTAKIRP-DVPVQVTIVPNADRTFKFSLRTPSSTW 82
P ++ Q +GPLG++ ++ TA + V VQ+ I DR ++ PS
Sbjct 24 PGATLAQRVGPLGLSSKVVGEDIKKATADYKSLKVHVQLAI---KDRKATVEVQ-PSVAT 79
Query 83 FLLRAARCPMGSEQAGKEVIGNVTLK--EIYHIAK 115
++++ + P + K ++ N +L+ E+ IA+
Sbjct 80 LIIKSLKEPPRDRKKEKNILHNGSLRMTEVVDIAR 114
> YML112w
Length=296
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 31/61 (50%), Gaps = 8/61 (13%)
Query 32 LGPLGINMMAFCKEFNARTAKIRPDV---PVQVTIVPN---ADRTFKFSLRTPSSTWFLL 85
L + +AF ++FN+ T I PD+ P I+ + ADR + R+ ++W++
Sbjct 192 LKDRDLKKLAFFQQFNSDTTAINPDLQTQPTNANILLHRMEADR--ELHKRSKETSWYIE 249
Query 86 R 86
R
Sbjct 250 R 250
> 7293815
Length=481
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 86 RAARCPMGSEQAGKEVIGNVTLKEI 110
R +CP G Q KEV+ VTL EI
Sbjct 219 RFVQCPEGELQKRKEVVHTVTLHEI 243
> Hs14751164
Length=550
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 35 LGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRTFKFSLRTPSSTWF 83
+GI K + K+ P+ P Q+ +P + + K SL TP S F
Sbjct 165 IGIGPPTLSKPTQTKGLKVEPEEPSQMPPLPQSHQKLKESLMTPGSGAF 213
> Hs4827067
Length=1370
Score = 27.3 bits (59), Expect = 7.2, Method: Composition-based stats.
Identities = 13/20 (65%), Positives = 16/20 (80%), Gaps = 1/20 (5%)
Query 19 AANAKPSPSIGQTLGPLGIN 38
+ NA PSPS+G+TLG GIN
Sbjct 186 SPNAPPSPSVGETLGD-GIN 204
> Hs20555958
Length=220
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 29/61 (47%), Gaps = 12/61 (19%)
Query 18 PAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRTFKFSLRT 77
P +KP P GQ+LGP GI+ M E+ +R PV +++ D + S +
Sbjct 136 PRKASKPLP--GQSLGPQGISAM----EYYSRKR------PVSSSVILQGDMNMRASAKK 183
Query 78 P 78
P
Sbjct 184 P 184
> At4g05040
Length=591
Score = 27.3 bits (59), Expect = 8.1, Method: Composition-based stats.
Identities = 9/27 (33%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 87 AARCPMGSEQAGKEVIGNVTLKEIYHI 113
A PMG ++G E + N+ L +++H+
Sbjct 67 GATPPMGDNESGLEFLNNLKLSDLFHL 93
> Hs20554894
Length=93
Score = 26.9 bits (58), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 79 SSTWFLLRAARCPMGSEQAGKEVIGNVTLKEI 110
STW+LL AA +GSE G + V + EI
Sbjct 17 ESTWWLLAAAGVEVGSELDGSFMFQQVPMVEI 48
Lambda K H
0.322 0.134 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40