bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2474_orf3
Length=135
Score E
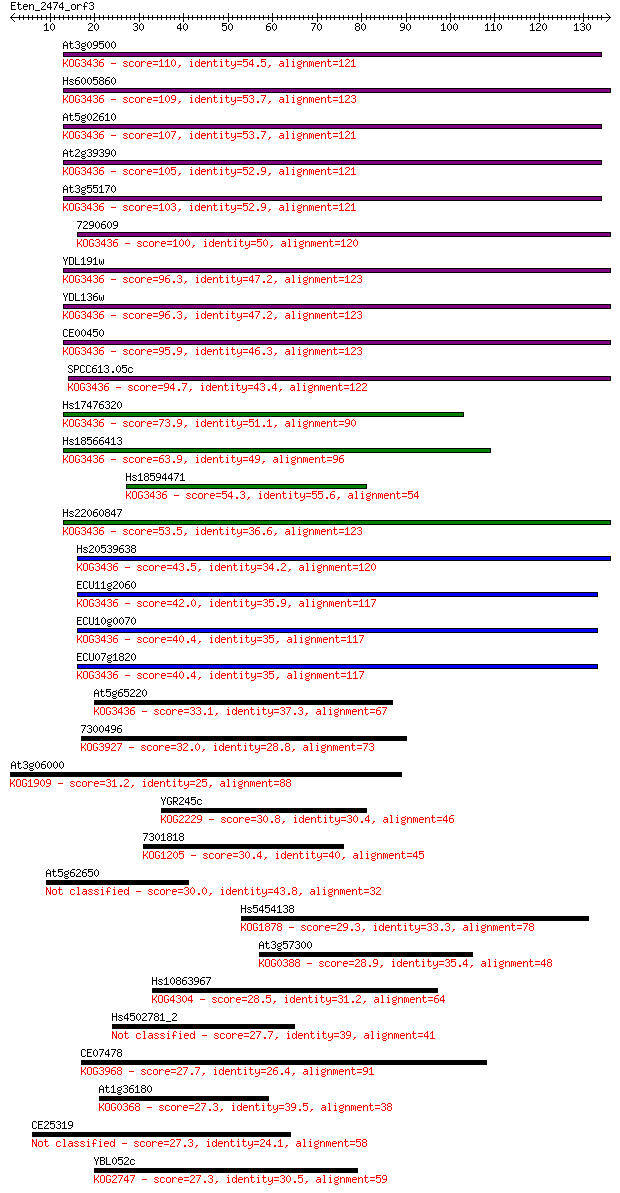
Sequences producing significant alignments: (Bits) Value
At3g09500 110 1e-24
Hs6005860 109 1e-24
At5g02610 107 5e-24
At2g39390 105 2e-23
At3g55170 103 7e-23
7290609 100 1e-21
YDL191w 96.3 2e-20
YDL136w 96.3 2e-20
CE00450 95.9 2e-20
SPCC613.05c 94.7 5e-20
Hs17476320 73.9 8e-14
Hs18566413 63.9 8e-11
Hs18594471 54.3 7e-08
Hs22060847 53.5 1e-07
Hs20539638 43.5 1e-04
ECU11g2060 42.0 3e-04
ECU10g0070 40.4 8e-04
ECU07g1820 40.4 8e-04
At5g65220 33.1 0.16
7300496 32.0 0.35
At3g06000 31.2 0.52
YGR245c 30.8 0.73
7301818 30.4 0.91
At5g62650 30.0 1.3
Hs5454138 29.3 2.3
At3g57300 28.9 2.6
Hs10863967 28.5 4.2
Hs4502781_2 27.7 5.7
CE07478 27.7 6.1
At1g36180 27.3 7.4
CE25319 27.3 7.9
YBL052c 27.3 8.6
> At3g09500
Length=123
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 66/121 (54%), Positives = 82/121 (67%), Gaps = 0/121 (0%)
Query 13 MVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTV 72
M I HELR+K+ DLQ QL++LK EL+LLRVAKVTG P KL+KI VRK IA+VLTV
Sbjct 1 MARIKVHELREKSKSDLQNQLKELKAELALLRVAKVTGGAPNKLSKIKVVRKSIAQVLTV 60
Query 73 YNQKRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
+QK++ ++ +K K P DLR KKTRAIR+RLT Q T R K+ P RKYA
Sbjct 61 SSQKQKSALREAYKNKKLLPLDLRPKKTRAIRRRLTKHQASLKTEREKKKEMYFPIRKYA 120
Query 133 L 133
+
Sbjct 121 I 121
> Hs6005860
Length=123
Score = 109 bits (273), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 66/123 (53%), Positives = 86/123 (69%), Gaps = 0/123 (0%)
Query 13 MVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTV 72
M I A +LR K ++L KQL+DLK ELS LRVAKVTG +KL+KI VRK IARVLTV
Sbjct 1 MAKIKARDLRGKKKEELLKQLDDLKVELSQLRVAKVTGGAASKLSKIRVVRKSIARVLTV 60
Query 73 YNQKRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
NQ ++E +KF+KG K+KP DLR KKTRA+R+RL + T ++ ++ + P RKYA
Sbjct 61 INQTQKENLRKFYKGKKYKPLDLRPKKTRAMRRRLNKHEENLKTKKQQRKERLYPLRKYA 120
Query 133 LLA 135
+ A
Sbjct 121 VKA 123
> At5g02610
Length=123
Score = 107 bits (268), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 65/121 (53%), Positives = 80/121 (66%), Gaps = 0/121 (0%)
Query 13 MVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTV 72
M I HELR K+ DLQ QL++ K EL+LLRVAKVTG P KL+KI VRK IA+VLTV
Sbjct 1 MARIKVHELRDKSKTDLQNQLKEFKAELALLRVAKVTGGAPNKLSKIKVVRKSIAQVLTV 60
Query 73 YNQKRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
+QK++ ++ +K K P DLR KKTRAIR+RLT Q T R K+ P RKYA
Sbjct 61 ISQKQKSALREAYKNKKLLPLDLRPKKTRAIRRRLTKHQASLKTEREKKKEMYFPVRKYA 120
Query 133 L 133
+
Sbjct 121 I 121
> At2g39390
Length=123
Score = 105 bits (263), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 64/121 (52%), Positives = 80/121 (66%), Gaps = 0/121 (0%)
Query 13 MVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTV 72
M I HELR+K+ DL QL++ K EL+LLRVAKVTG P KL+KI VRK IA+VLTV
Sbjct 1 MARIKVHELREKSKADLSGQLKEFKAELALLRVAKVTGGAPNKLSKIKVVRKSIAQVLTV 60
Query 73 YNQKRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
+QK++ ++ +K K P DLR KKTRAIR+RLT Q T R K+ P RKYA
Sbjct 61 ISQKQKSALREAYKNKKLLPLDLRPKKTRAIRRRLTKHQASLKTEREKKKEMYFPIRKYA 120
Query 133 L 133
+
Sbjct 121 I 121
> At3g55170
Length=123
Score = 103 bits (258), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 64/121 (52%), Positives = 79/121 (65%), Gaps = 0/121 (0%)
Query 13 MVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTV 72
M I HELR K+ DL QL++LK EL+ LRVAKVTG P KL+KI VRK IA+VLTV
Sbjct 1 MARIKVHELRDKSKSDLSTQLKELKAELASLRVAKVTGGAPNKLSKIKVVRKSIAQVLTV 60
Query 73 YNQKRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
+QK++ ++ +K K P DLR KKTRAIR+RLT Q T R K+ P RKYA
Sbjct 61 SSQKQKSALREAYKNKKLLPLDLRPKKTRAIRRRLTKHQASLKTEREKKKDMYFPIRKYA 120
Query 133 L 133
+
Sbjct 121 I 121
> 7290609
Length=121
Score = 100 bits (248), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 60/120 (50%), Positives = 81/120 (67%), Gaps = 0/120 (0%)
Query 16 ITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTVYNQ 75
+ ELR K K+L KQL++LK EL LRVAKVTG P+KL+KI VRK IARV V +Q
Sbjct 2 VKCSELRIKDKKELTKQLDELKNELLSLRVAKVTGGAPSKLSKIRVVRKAIARVYIVMHQ 61
Query 76 KRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYALLA 135
K++E +K FK K+KP DLR KKTRAIR+ L+ + T++ ++ P+RK+A+ A
Sbjct 62 KQKENLRKVFKNKKYKPLDLRKKKTRAIRKALSPRDANRKTLKEIRKRSVFPQRKFAVKA 121
> YDL191w
Length=120
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 58/123 (47%), Positives = 85/123 (69%), Gaps = 3/123 (2%)
Query 13 MVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTV 72
M G+ A+ELR K+ + L QL DLKKEL+ L+V K++ + L KI VRK IA VLTV
Sbjct 1 MAGVKAYELRTKSKEQLASQLVDLKKELAELKVQKLSRPS---LPKIKTVRKSIACVLTV 57
Query 73 YNQKRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
N+++RE ++ +KG K++PKDLR KKTRA+R+ LT + ++T ++ K+ P+RKYA
Sbjct 58 INEQQREAVRQLYKGKKYQPKDLRAKKTRALRRALTKFEASQVTEKQRKKQIAFPQRKYA 117
Query 133 LLA 135
+ A
Sbjct 118 IKA 120
> YDL136w
Length=120
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 58/123 (47%), Positives = 85/123 (69%), Gaps = 3/123 (2%)
Query 13 MVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTV 72
M G+ A+ELR K+ + L QL DLKKEL+ L+V K++ + L KI VRK IA VLTV
Sbjct 1 MAGVKAYELRTKSKEQLASQLVDLKKELAELKVQKLSRPS---LPKIKTVRKSIACVLTV 57
Query 73 YNQKRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
N+++RE ++ +KG K++PKDLR KKTRA+R+ LT + ++T ++ K+ P+RKYA
Sbjct 58 INEQQREAVRQLYKGKKYQPKDLRAKKTRALRRALTKFEASQVTEKQRKKQIAFPQRKYA 117
Query 133 LLA 135
+ A
Sbjct 118 IKA 120
> CE00450
Length=123
Score = 95.9 bits (237), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 57/123 (46%), Positives = 82/123 (66%), Gaps = 0/123 (0%)
Query 13 MVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTV 72
M + LR + LQK+L++ K EL+ LRV+KVTG +KL+KI VRK IAR+LTV
Sbjct 1 MTKLKCKSLRGEKKDALQKKLDEQKTELATLRVSKVTGGAASKLSKIRVVRKNIARLLTV 60
Query 73 YNQKRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
NQ +++E +KF+ K+KP DLRLKKTRAIR+RLT + + ++ + +N RK+A
Sbjct 61 INQTQKQELRKFYADHKYKPIDLRLKKTRAIRRRLTAHELSLRSAKQQAKSRNQAVRKFA 120
Query 133 LLA 135
+ A
Sbjct 121 VKA 123
> SPCC613.05c
Length=122
Score = 94.7 bits (234), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 53/122 (43%), Positives = 85/122 (69%), Gaps = 0/122 (0%)
Query 14 VGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTVY 73
+ + ELR+++ ++L +QL++L++EL+ LRV K+ G + +KL+KI RK IAR+LTV
Sbjct 1 MALKTFELRKQSQENLAEQLQELRQELASLRVQKIAGGSGSKLSKIKTTRKDIARILTVI 60
Query 74 NQKRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYAL 133
N+ R A++ +K K+ P DLR KKTRAIR+ LT ++ + T+++ K+ + P RKYAL
Sbjct 61 NESNRLAAREAYKNKKYIPLDLRQKKTRAIRRALTPYEQSRKTLKQIKKERYFPLRKYAL 120
Query 134 LA 135
A
Sbjct 121 KA 122
> Hs17476320
Length=101
Score = 73.9 bits (180), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 46/90 (51%), Positives = 59/90 (65%), Gaps = 0/90 (0%)
Query 13 MVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTV 72
M I + +L +K +L KQL DLK E S LRVAKVTG +KL+KI V K +ARVLTV
Sbjct 1 MAKIKSRDLHRKKEAELLKQLGDLKVERSQLRVAKVTGGEASKLSKIRVVCKSVARVLTV 60
Query 73 YNQKRREEAKKFFKGCKHKPKDLRLKKTRA 102
NQ ++E +KF KG + +P LR +KT A
Sbjct 61 INQTQKENIRKFCKGKRCEPLTLRPEKTHA 90
> Hs18566413
Length=99
Score = 63.9 bits (154), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 47/96 (48%), Positives = 56/96 (58%), Gaps = 19/96 (19%)
Query 13 MVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTV 72
M I A +LR K ++L KQL+DLK ELS LRVAKVTG +KL KI VRK IA VLTV
Sbjct 1 MAKIKAPDLRGK-KEELLKQLDDLKVELSQLRVAKVTGGAASKLPKILVVRKSIAHVLTV 59
Query 73 YNQKRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLT 108
NQ ++ KKTRA+R+RL
Sbjct 60 INQTQKA------------------KKTRAMRRRLN 77
> Hs18594471
Length=94
Score = 54.3 bits (129), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 30/54 (55%), Positives = 40/54 (74%), Gaps = 0/54 (0%)
Query 27 KDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTVYNQKRREE 80
++L KQL++LK ELS LRV KVTG ++L+KI VRK IA VL V NQ++ +E
Sbjct 27 EELLKQLDNLKVELSQLRVTKVTGGAASRLSKIRVVRKSIACVLMVINQRKPQE 80
> Hs22060847
Length=94
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 45/123 (36%), Positives = 63/123 (51%), Gaps = 29/123 (23%)
Query 13 MVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTV 72
M I A +LR K ++L KQL+DLK ELS LRVAK
Sbjct 1 MAKIKARDLRGKK-EELLKQLDDLKVELSQLRVAKT------------------------ 35
Query 73 YNQKRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
++E +KF+KG +KP DLR KKTRA+R+RL + T ++ ++ + P KYA
Sbjct 36 ----QKENLRKFYKGRNYKPLDLRPKKTRAMRRRLNKHEENPKTKKQQRKERLYPLGKYA 91
Query 133 LLA 135
+ A
Sbjct 92 VKA 94
> Hs20539638
Length=219
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 41/120 (34%), Positives = 59/120 (49%), Gaps = 30/120 (25%)
Query 16 ITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTVYNQ 75
I A +L +K ++L KQL+DLK ELS L VAK
Sbjct 124 IKARDLHEK--EELLKQLDDLKVELSQLHVAKT--------------------------- 154
Query 76 KRREEAKKFFKGCKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYALLA 135
++E+ +KF+KG K+KP DL KKTRA+ R + T +R ++ + RKYA+ A
Sbjct 155 -QKEKLRKFYKGKKYKPLDLWPKKTRAMCCRFNKHKENLKTKKRQRKERLCLLRKYAVKA 213
> ECU11g2060
Length=122
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 42/119 (35%), Positives = 64/119 (53%), Gaps = 4/119 (3%)
Query 16 ITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTVYNQ 75
I A LRQ K ++++ ++K EL+ LR K +G A I +K +AR LTV +
Sbjct 3 IEASALRQLGIKQIEERAAEIKAELAALRQKKNSGDVGAN--DIKTAKKNLARALTVRRE 60
Query 76 KRREEAKKFFKG--CKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
K EE + ++G PK+LR K R+ R+ LT +Q R+ T R+ R+ PR +A
Sbjct 61 KILEELVEAYRGTPVSKLPKELRPKLNRSKRRALTKTQLRRKTRRQRARMSKFPRVIFA 119
> ECU10g0070
Length=122
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 41/119 (34%), Positives = 64/119 (53%), Gaps = 4/119 (3%)
Query 16 ITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTVYNQ 75
I A LRQ K ++++ ++K +L+ LR K +G A I +K +AR LTV +
Sbjct 3 IEASALRQLGIKQIEERAAEIKADLAALRQKKNSGDVGAN--DIKTAKKNLARALTVRRE 60
Query 76 KRREEAKKFFKG--CKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
K EE + ++G PK+LR K R+ R+ LT +Q R+ T R+ R+ PR +A
Sbjct 61 KILEELVEAYRGTPVSKLPKELRPKLNRSKRRALTKTQLRRKTRRQRARMSKFPRVIFA 119
> ECU07g1820
Length=122
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 41/119 (34%), Positives = 64/119 (53%), Gaps = 4/119 (3%)
Query 16 ITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTVYNQ 75
I A LRQ K ++++ ++K +L+ LR K +G A I +K +AR LTV +
Sbjct 3 IEASALRQLGIKQIEERAAEIKADLAALRQKKNSGDVGAN--DIKTAKKNLARALTVRRE 60
Query 76 KRREEAKKFFKG--CKHKPKDLRLKKTRAIRQRLTLSQRRKMTVRRTKRVQNVPRRKYA 132
K EE + ++G PK+LR K R+ R+ LT +Q R+ T R+ R+ PR +A
Sbjct 61 KILEELVEAYRGTPVSKLPKELRPKLNRSKRRALTKTQLRRKTRRQRARMSKFPRVIFA 119
> At5g65220
Length=173
Score = 33.1 bits (74), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 40/67 (59%), Gaps = 4/67 (5%)
Query 20 ELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTVYNQKRRE 79
E+R K + LQ+++ DLK EL +LR+ K + K + +++K +AR+LTV KR
Sbjct 69 EIRSKTTEQLQEEVVDLKGELFMLRLQK-SARNEFKSSDFRRMKKQVARMLTV---KRER 124
Query 80 EAKKFFK 86
E K+ K
Sbjct 125 EIKEGIK 131
> 7300496
Length=353
Score = 32.0 bits (71), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 34/80 (42%), Gaps = 8/80 (10%)
Query 17 TAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGAT-------PAKLAKITQVRKGIARV 69
T +L ++AP LQ + L + +VT P + ++ +G+ RV
Sbjct 174 TPDDLPKEAPASLQDTVGKLGNTPKIWITCEVTNGPKPEMVFYPGPYFEASENMRGVTRV 233
Query 70 LTVYNQKRREEAKKFFKGCK 89
+ + K E AK FF CK
Sbjct 234 VAIQMNKMPENAKTFFS-CK 252
> At3g06000
Length=211
Score = 31.2 bits (69), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 45/92 (48%), Gaps = 5/92 (5%)
Query 1 ISACLGSSPHIKMVGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKIT 60
I+ CL + H+K + ++ ++L+ + ++ K +ED + E + + +LA++
Sbjct 86 IAVCLAAKRHLKKLNLSENDLKDEGCVEIVKSMEDWELEYVDMSYNDLRREGALRLARVV 145
Query 61 QVRKGIARVLTVYNQ----KRREEAKKFFKGC 88
V+KG ++L + K EE K F C
Sbjct 146 -VKKGSFKMLNIDGNMISLKGIEEIKVIFTNC 176
> YGR245c
Length=767
Score = 30.8 bits (68), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 28/48 (58%), Gaps = 2/48 (4%)
Query 35 DLKKELSLLRVAKVTGATPAKLAKITQVR--KGIARVLTVYNQKRREE 80
D+ E + +A TPA AK+ ++R + +A+++ ++ Q +REE
Sbjct 626 DIDPEAAFREIASTRILTPADFAKLQELRNEESVAKIMGIHKQDKREE 673
> 7301818
Length=326
Score = 30.4 bits (67), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 27/47 (57%), Gaps = 2/47 (4%)
Query 31 KQLEDLKKELSLLRVAKVTGAT--PAKLAKITQVRKGIARVLTVYNQ 75
++LE +KK+L L V T P LA++ + + + RVL VYNQ
Sbjct 88 QELERVKKDLLALDVDPAYPPTVLPLDLAELNSIPEFVTRVLAVYNQ 134
> At5g62650
Length=451
Score = 30.0 bits (66), Expect = 1.3, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 9 PHIKMVGITAHELRQKAPKDLQKQLEDLKKEL 40
P I MVGI+ E Q + +L+K +EDL ++L
Sbjct 368 PKITMVGISTGEAAQMSKANLKKTMEDLTEDL 399
> Hs5454138
Length=2440
Score = 29.3 bits (64), Expect = 2.3, Method: Composition-based stats.
Identities = 26/111 (23%), Positives = 46/111 (41%), Gaps = 33/111 (29%)
Query 53 PAKLAKITQVRKGIARVLTVYNQKRREEAKKFFKGCKHK--------PKDLR-----LKK 99
P + Q + I +++ N+K+ EEA K F+G K P D + +K
Sbjct 222 PVSPPPVEQKHRSIVQIIYDENRKKAEEAHKIFEGLGPKVELPLYNQPSDTKVYHENIKT 281
Query 100 TRAIRQRLTLSQRRKMTVRRTK--------------------RVQNVPRRK 130
+ +R++L L +R+ R+ + R++N PRRK
Sbjct 282 NQVMRKKLILFFKRRNHARKQREQKICQRYDQLMEAWEKKVDRIENNPRRK 332
> At3g57300
Length=1496
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 31/51 (60%), Gaps = 6/51 (11%)
Query 57 AKITQVRKGIA---RVLTVYNQKRREEAKKFFKGCKHKPKDLRLKKTRAIR 104
A I VR+ IA R+ T +++K +AK+F GC+ +++R+K R+ +
Sbjct 350 AWINIVRRDIAKHHRIFTTFHRKLSIDAKRFADGCQ---REVRMKVGRSYK 397
> Hs10863967
Length=221
Score = 28.5 bits (62), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 27/64 (42%), Gaps = 8/64 (12%)
Query 33 LEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTVYNQKRREEAKKFFKGCKHKP 92
LE + L LR +VT A A A + + R G L E +F GC+ P
Sbjct 81 LEMTVRHLRSLRRVQVTAALSADPAVLGKYRAGFHECLA--------EVNRFLAGCEGVP 132
Query 93 KDLR 96
D+R
Sbjct 133 ADVR 136
> Hs4502781_2
Length=1932
Score = 27.7 bits (60), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 29/42 (69%), Gaps = 1/42 (2%)
Query 24 KAPKDLQKQLEDL-KKELSLLRVAKVTGATPAKLAKITQVRK 64
K+ +LQK++++L KKEL LLRV + + K+ ++ Q++K
Sbjct 1232 KSKDELQKKIQELQKKELQLLRVKEDVNMSHKKINEMEQLKK 1273
> CE07478
Length=445
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 24/92 (26%), Positives = 38/92 (41%), Gaps = 3/92 (3%)
Query 17 TAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTVYNQK 76
T ++ R+ + +DL+K E+L K+ +LR T + + RVL N
Sbjct 128 TTNKTREASEQDLRKDFEELAKK--MLRSGTTTLEAKSGYGLNVDAEMKMLRVLATENPN 185
Query 77 RREEAKKFFKGCKHKPK-DLRLKKTRAIRQRL 107
E F G PK ++TR I + L
Sbjct 186 IPLEVSATFCGAHAVPKGSTEYEQTRMICEEL 217
> At1g36180
Length=2359
Score = 27.3 bits (59), Expect = 7.4, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 21 LRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAK 58
L + PKDL+ LE KE ++ +T PAKL K
Sbjct 937 LATRLPKDLRNMLELKYKEFEIISKTSLTPDFPAKLLK 974
> CE25319
Length=902
Score = 27.3 bits (59), Expect = 7.9, Method: Composition-based stats.
Identities = 14/59 (23%), Positives = 28/59 (47%), Gaps = 1/59 (1%)
Query 6 GSSPHIKM-VGITAHELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVR 63
G P ++M + +R + P D ++D KE +++ ATP+K+ ++ R
Sbjct 46 GQPPDLEMNLPSKQRRVRTRKPMDTDGSIDDGDKEKKAMKIINPAAATPSKMFRVQGER 104
> YBL052c
Length=831
Score = 27.3 bits (59), Expect = 8.6, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 30/59 (50%), Gaps = 6/59 (10%)
Query 20 ELRQKAPKDLQKQLEDLKKELSLLRVAKVTGATPAKLAKITQVRKGIARVLTVYNQKRR 78
+LR A + + ED +++SL +AK+TG P T V G+ ++ +Y K R
Sbjct 477 KLRDSARRRSNNKNEDTFQQVSLNDIAKLTGMIP------TDVVFGLEQLQVLYRHKTR 529
Lambda K H
0.321 0.132 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1388795348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40