bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2466_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
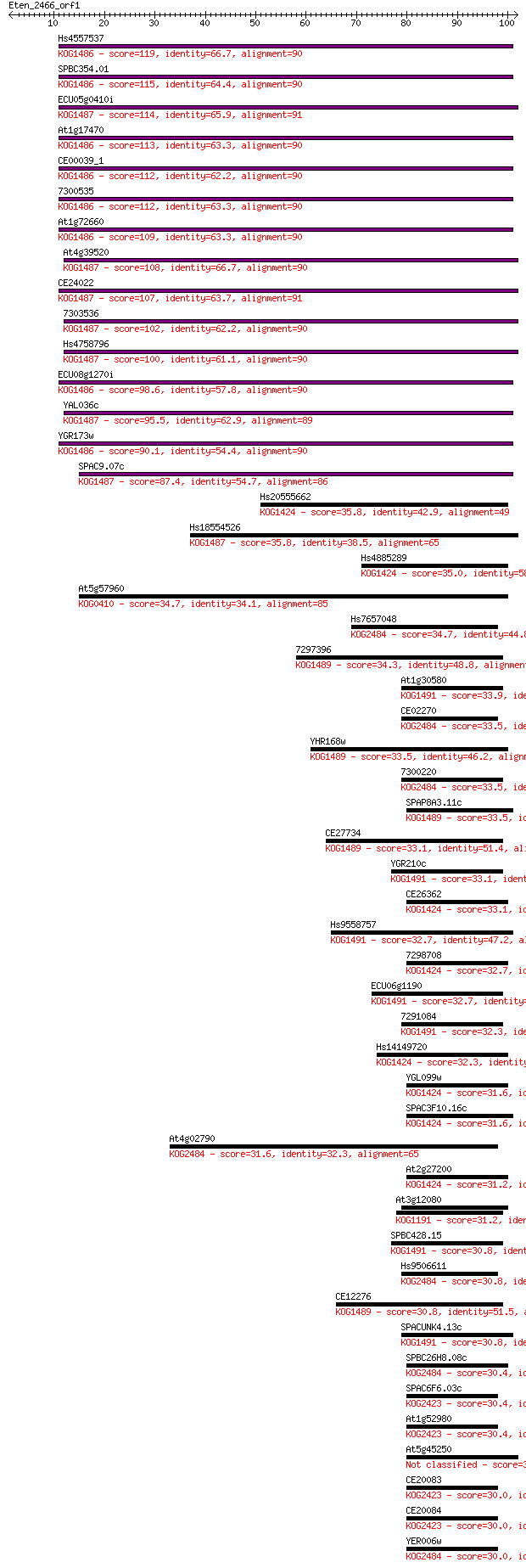
Hs4557537 119 1e-27
SPBC354.01 115 2e-26
ECU05g0410i 114 4e-26
At1g17470 113 1e-25
CE00039_1 112 1e-25
7300535 112 2e-25
At1g72660 109 1e-24
At4g39520 108 2e-24
CE24022 107 4e-24
7303536 102 1e-22
Hs4758796 100 7e-22
ECU08g1270i 98.6 3e-21
YAL036c 95.5 2e-20
YGR173w 90.1 9e-19
SPAC9.07c 87.4 6e-18
Hs20555662 35.8 0.018
Hs18554526 35.8 0.020
Hs4885289 35.0 0.031
At5g57960 34.7 0.044
Hs7657048 34.7 0.045
7297396 34.3 0.056
At1g30580 33.9 0.077
CE02270 33.5 0.088
YHR168w 33.5 0.10
7300220 33.5 0.10
SPAP8A3.11c 33.5 0.11
CE27734 33.1 0.12
YGR210c 33.1 0.13
CE26362 33.1 0.14
Hs9558757 32.7 0.15
7298708 32.7 0.16
ECU06g1190 32.7 0.16
7291084 32.3 0.21
Hs14149720 32.3 0.24
YGL099w 31.6 0.36
SPAC3F10.16c 31.6 0.36
At4g02790 31.6 0.36
At2g27200 31.2 0.43
At3g12080 31.2 0.49
SPBC428.15 30.8 0.57
Hs9506611 30.8 0.61
CE12276 30.8 0.63
SPACUNK4.13c 30.8 0.64
SPBC26H8.08c 30.4 0.79
SPAC6F6.03c 30.4 0.80
At1g52980 30.4 0.91
At5g45250 30.0 1.0
CE20083 30.0 1.1
CE20084 30.0 1.1
YER006w 30.0 1.1
> Hs4557537
Length=364
Score = 119 bits (299), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 60/90 (66%), Positives = 71/90 (78%), Gaps = 5/90 (5%)
Query 11 MSVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGF 70
M +L+KISE+E EIARTQKNKAT +HLGLLKAKLAK R QLLE S S+SS GEGF
Sbjct 1 MGILEKISEIEKEIARTQKNKATEYHLGLLKAKLAKYRAQLLEPSKSASSK-----GEGF 55
Query 71 EVSKTGDPRIGLVGFPSVGKSTLLNKLTGT 100
+V K+GD R+ L+GFPSVGKST L+ +T T
Sbjct 56 DVMKSGDARVALIGFPSVGKSTFLSLMTST 85
> SPBC354.01
Length=346
Score = 115 bits (289), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 58/90 (64%), Positives = 70/90 (77%), Gaps = 5/90 (5%)
Query 11 MSVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGF 70
M VL+KI E+EAE+ RTQKNKAT +HLGLLK KLAKLR QLLE +S+SG GEGF
Sbjct 1 MGVLEKIQEIEAEMRRTQKNKATEYHLGLLKGKLAKLRAQLLE-----PTSKSGPKGEGF 55
Query 71 EVSKTGDPRIGLVGFPSVGKSTLLNKLTGT 100
+V K+GD R+ +GFPSVGKSTLL+ +T T
Sbjct 56 DVLKSGDARVAFIGFPSVGKSTLLSAITKT 85
> ECU05g0410i
Length=362
Score = 114 bits (285), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 60/92 (65%), Positives = 76/92 (82%), Gaps = 7/92 (7%)
Query 11 MSVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGE-G 69
M+V++KI EVE+E+ARTQKNKAT+ HLG+LKAKLAKLR +L+ S+ +SG GE G
Sbjct 1 MNVMEKIKEVESEMARTQKNKATSNHLGILKAKLAKLRRELI------SAPKSGSSGEGG 54
Query 70 FEVSKTGDPRIGLVGFPSVGKSTLLNKLTGTF 101
F+V+KTG R+G VGFPSVGKSTL++KLTGTF
Sbjct 55 FDVAKTGIARVGFVGFPSVGKSTLMSKLTGTF 86
> At1g17470
Length=399
Score = 113 bits (282), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 57/90 (63%), Positives = 69/90 (76%), Gaps = 5/90 (5%)
Query 11 MSVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGF 70
M ++++I E+EAE+ARTQKNKAT +HLG LKAK+AKLR QLLE +S GGEGF
Sbjct 1 MGIIERIKEIEAEMARTQKNKATEYHLGQLKAKIAKLRTQLLEPPKGASG-----GGEGF 55
Query 71 EVSKTGDPRIGLVGFPSVGKSTLLNKLTGT 100
EV+K G R+ L+GFPSVGKSTLL LTGT
Sbjct 56 EVTKYGHGRVALIGFPSVGKSTLLTMLTGT 85
> CE00039_1
Length=408
Score = 112 bits (281), Expect = 1e-25, Method: Composition-based stats.
Identities = 56/90 (62%), Positives = 70/90 (77%), Gaps = 5/90 (5%)
Query 11 MSVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGF 70
M +L+KI+E+E EI+RTQKNKAT +HLGLLKAKLAK R QLLE + + G GEGF
Sbjct 1 MGILEKIAEIEHEISRTQKNKATEYHLGLLKAKLAKYRQQLLE-----PTGKGGAKGEGF 55
Query 71 EVSKTGDPRIGLVGFPSVGKSTLLNKLTGT 100
+V K+GD R+ +VGFPSVGKSTLL+ +T T
Sbjct 56 DVMKSGDARVAMVGFPSVGKSTLLSSMTST 85
> 7300535
Length=363
Score = 112 bits (280), Expect = 2e-25, Method: Composition-based stats.
Identities = 57/90 (63%), Positives = 70/90 (77%), Gaps = 6/90 (6%)
Query 11 MSVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGF 70
M +L+KI+E+E EIARTQKNKAT +HLGLLKAKLAK R QLLE S+ G G+GF
Sbjct 1 MGILEKIAEIEREIARTQKNKATEYHLGLLKAKLAKYRSQLLE------PSKKGEKGDGF 54
Query 71 EVSKTGDPRIGLVGFPSVGKSTLLNKLTGT 100
+V K+GD R+ L+GFPSVGKST+L+ LT T
Sbjct 55 DVLKSGDARVALIGFPSVGKSTMLSTLTKT 84
> At1g72660
Length=399
Score = 109 bits (273), Expect = 1e-24, Method: Composition-based stats.
Identities = 57/90 (63%), Positives = 69/90 (76%), Gaps = 5/90 (5%)
Query 11 MSVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGF 70
M ++++I E+EAE+ARTQKNKAT +HLG LKAK+AKLR QLLE SS GG+GF
Sbjct 1 MGIVERIKEIEAEMARTQKNKATEYHLGQLKAKIAKLRTQLLEPPKGSSG-----GGDGF 55
Query 71 EVSKTGDPRIGLVGFPSVGKSTLLNKLTGT 100
EV+K G R+ L+GFPSVGKSTLL LTGT
Sbjct 56 EVTKYGHGRVALIGFPSVGKSTLLTMLTGT 85
> At4g39520
Length=369
Score = 108 bits (271), Expect = 2e-24, Method: Composition-based stats.
Identities = 60/90 (66%), Positives = 73/90 (81%), Gaps = 3/90 (3%)
Query 12 SVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGFE 71
+++QKI E+E E+A+TQKNKAT+ HLGLLKAKLAKLR LL + + G GEGF+
Sbjct 3 TIMQKIKEIEDEMAKTQKNKATSHHLGLLKAKLAKLRRDLL---APPTKGGGGGAGEGFD 59
Query 72 VSKTGDPRIGLVGFPSVGKSTLLNKLTGTF 101
V+K+GD R+GLVGFPSVGKSTLLNKLTGTF
Sbjct 60 VTKSGDSRVGLVGFPSVGKSTLLNKLTGTF 89
> CE24022
Length=366
Score = 107 bits (268), Expect = 4e-24, Method: Composition-based stats.
Identities = 58/91 (63%), Positives = 66/91 (72%), Gaps = 5/91 (5%)
Query 11 MSVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGF 70
MSVLQKI+++EAE+ARTQKNKATN HLG+LKAKLAKLR L+ GF
Sbjct 1 MSVLQKIADIEAEMARTQKNKATNAHLGILKAKLAKLRRDLITPKGGGGGPGE-----GF 55
Query 71 EVSKTGDPRIGLVGFPSVGKSTLLNKLTGTF 101
+V+KTGD RIG VGFPSVGKSTLL L G F
Sbjct 56 DVAKTGDARIGFVGFPSVGKSTLLCNLAGVF 86
> 7303536
Length=368
Score = 102 bits (255), Expect = 1e-22, Method: Composition-based stats.
Identities = 56/90 (62%), Positives = 69/90 (76%), Gaps = 4/90 (4%)
Query 12 SVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGFE 71
++L+KIS +E+E+ARTQKNKAT+ HLGLLKAKLAKLR +L+ S G G GFE
Sbjct 3 TILEKISAIESEMARTQKNKATSAHLGLLKAKLAKLRRELI----SPKGGGGGTGEAGFE 58
Query 72 VSKTGDPRIGLVGFPSVGKSTLLNKLTGTF 101
V+KTGD R+G VGFPSVGKSTLL+ L G +
Sbjct 59 VAKTGDARVGFVGFPSVGKSTLLSNLAGVY 88
> Hs4758796
Length=367
Score = 100 bits (249), Expect = 7e-22, Method: Composition-based stats.
Identities = 55/90 (61%), Positives = 65/90 (72%), Gaps = 4/90 (4%)
Query 12 SVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGFE 71
S L KI+E+EAE+ARTQKNKAT HLGLLKA+LAKLR +L+ EGF+
Sbjct 3 STLAKIAEIEAEMARTQKNKATAHHLGLLKARLAKLRRELITPKGGGGGGPG----EGFD 58
Query 72 VSKTGDPRIGLVGFPSVGKSTLLNKLTGTF 101
V+KTGD RIG VGFPSVGKSTLL+ L G +
Sbjct 59 VAKTGDARIGFVGFPSVGKSTLLSNLAGVY 88
> ECU08g1270i
Length=362
Score = 98.6 bits (244), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 52/90 (57%), Positives = 65/90 (72%), Gaps = 6/90 (6%)
Query 11 MSVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGF 70
M + KI+E+EAE+ARTQKNK T HLG LKAKLA+ R +L + SS S +GF
Sbjct 1 MGIQDKINEIEAEMARTQKNKKTEHHLGALKAKLARYRQELDVPKTRSSKS------DGF 54
Query 71 EVSKTGDPRIGLVGFPSVGKSTLLNKLTGT 100
EVSK+GD R+ L+G PSVGKSTLL+K+T T
Sbjct 55 EVSKSGDARVVLIGLPSVGKSTLLSKITST 84
> YAL036c
Length=369
Score = 95.5 bits (236), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 56/89 (62%), Positives = 70/89 (78%), Gaps = 3/89 (3%)
Query 12 SVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGFE 71
+ ++KI +E E+ARTQKNKAT+FHLG LKAKLAKLR +LL +S+SS G G GF+
Sbjct 3 TTVEKIKAIEDEMARTQKNKATSFHLGQLKAKLAKLRRELL---TSASSGSGGGAGIGFD 59
Query 72 VSKTGDPRIGLVGFPSVGKSTLLNKLTGT 100
V++TG +G VGFPSVGKSTLL+KLTGT
Sbjct 60 VARTGVASVGFVGFPSVGKSTLLSKLTGT 88
> YGR173w
Length=368
Score = 90.1 bits (222), Expect = 9e-19, Method: Composition-based stats.
Identities = 49/90 (54%), Positives = 62/90 (68%), Gaps = 4/90 (4%)
Query 11 MSVLQKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGF 70
M ++ KI +E E+ARTQKNKAT HLGLLK KLA+ R QLL + S GF
Sbjct 1 MGIIDKIKAIEEEMARTQKNKATEHHLGLLKGKLARYRQQLLADEAGSGGGGG----SGF 56
Query 71 EVSKTGDPRIGLVGFPSVGKSTLLNKLTGT 100
EV+K+GD R+ L+G+PSVGKS+LL K+T T
Sbjct 57 EVAKSGDARVVLIGYPSVGKSSLLGKITTT 86
> SPAC9.07c
Length=366
Score = 87.4 bits (215), Expect = 6e-18, Method: Composition-based stats.
Identities = 47/86 (54%), Positives = 61/86 (70%), Gaps = 5/86 (5%)
Query 15 QKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGFEVSK 74
QKI EVE E+A+TQKNKAT HLG+LKAKLAKL+ +L+ + GF+V++
Sbjct 6 QKIKEVEDEMAKTQKNKATAKHLGMLKAKLAKLKRELITPTGGGGGGGL-----GFDVAR 60
Query 75 TGDPRIGLVGFPSVGKSTLLNKLTGT 100
TG +G +GFPSVGKSTL+ +LTGT
Sbjct 61 TGIGTVGFIGFPSVGKSTLMTQLTGT 86
> Hs20555662
Length=607
Score = 35.8 bits (81), Expect = 0.018, Method: Composition-based stats.
Identities = 21/49 (42%), Positives = 29/49 (59%), Gaps = 3/49 (6%)
Query 51 LLEGSSSSSSSRSGRGGEGFEVSKTGDPRIGLVGFPSVGKSTLLNKLTG 99
L+E + S+ +G E + K G IG VGFP+VGKS+L+N L G
Sbjct 337 LVEQQTDSAMEPTGPTQERY---KDGVVTIGCVGFPNVGKSSLINGLVG 382
> Hs18554526
Length=122
Score = 35.8 bits (81), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 33/68 (48%), Gaps = 3/68 (4%)
Query 37 LGLLKAKLAKLRGQLLEGSSSSSSSRSGR---GGEGFEVSKTGDPRIGLVGFPSVGKSTL 93
+G L ++ L + L G SS R+ R + +TGD IG V FPSVGKS
Sbjct 1 MGTLPRQVTGLDFKGLFGDFQVSSFRTSREFSHAKRLSFDRTGDAYIGSVAFPSVGKSVD 60
Query 94 LNKLTGTF 101
L L G +
Sbjct 61 LRNLAGLY 68
> Hs4885289
Length=430
Score = 35.0 bits (79), Expect = 0.031, Method: Composition-based stats.
Identities = 17/29 (58%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 71 EVSKTGDPRIGLVGFPSVGKSTLLNKLTG 99
E K G IG VGFP+VGKS+L+N L G
Sbjct 177 ERYKDGVVTIGCVGFPNVGKSSLINGLVG 205
> At5g57960
Length=540
Score = 34.7 bits (78), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 39/85 (45%), Gaps = 22/85 (25%)
Query 15 QKISEVEAEIARTQKNKATNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGFEVSK 74
+K EV+ I RTQ +G+LK +L +R RS R
Sbjct 269 EKQIEVDKRILRTQ--------IGVLKKELESVRKH-------RKQYRSRRVAIPV---- 309
Query 75 TGDPRIGLVGFPSVGKSTLLNKLTG 99
P + LVG+ + GKSTLLN+LTG
Sbjct 310 ---PVVSLVGYTNAGKSTLLNQLTG 331
> Hs7657048
Length=560
Score = 34.7 bits (78), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 69 GFEVSKTGDPRIGLVGFPSVGKSTLLNKL 97
GF+ + + R+G++GFP+VGKS+++N L
Sbjct 246 GFQETCSKAIRVGVIGFPNVGKSSIINSL 274
> 7297396
Length=381
Score = 34.3 bits (77), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 28/45 (62%), Gaps = 4/45 (8%)
Query 58 SSSSRSGRGGEGFEVSKTGDPR----IGLVGFPSVGKSTLLNKLT 98
+S S G G ++S T + R +GL+G+P+ GKSTLLN LT
Sbjct 182 TSPKVSEYGPRGEDLSYTLELRSMADVGLIGYPNAGKSTLLNALT 226
> At1g30580
Length=394
Score = 33.9 bits (76), Expect = 0.077, Method: Composition-based stats.
Identities = 14/20 (70%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 79 RIGLVGFPSVGKSTLLNKLT 98
+IG+VG P+VGKSTL N LT
Sbjct 26 KIGIVGLPNVGKSTLFNTLT 45
> CE02270
Length=556
Score = 33.5 bits (75), Expect = 0.088, Method: Compositional matrix adjust.
Identities = 12/19 (63%), Positives = 18/19 (94%), Gaps = 0/19 (0%)
Query 79 RIGLVGFPSVGKSTLLNKL 97
R+G+VGFP+VGKS+++N L
Sbjct 261 RVGVVGFPNVGKSSVINSL 279
> YHR168w
Length=499
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 26/43 (60%), Gaps = 6/43 (13%)
Query 61 SRSGRGGEG----FEVSKTGDPRIGLVGFPSVGKSTLLNKLTG 99
S+ GR G FE+ D +GL+G P+ GKST+LNK++
Sbjct 321 SKPGRNGLEQHFLFELKSIAD--LGLIGLPNAGKSTILNKISN 361
> 7300220
Length=581
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 12/20 (60%), Positives = 18/20 (90%), Gaps = 0/20 (0%)
Query 79 RIGLVGFPSVGKSTLLNKLT 98
R+G+VG P+VGKS+++N LT
Sbjct 269 RVGVVGIPNVGKSSIINSLT 288
> SPAP8A3.11c
Length=419
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 15/21 (71%), Positives = 17/21 (80%), Gaps = 0/21 (0%)
Query 80 IGLVGFPSVGKSTLLNKLTGT 100
IGLVG P+ GKSTLLN LT +
Sbjct 239 IGLVGLPNAGKSTLLNCLTAS 259
> CE27734
Length=390
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 18/37 (48%), Positives = 22/37 (59%), Gaps = 2/37 (5%)
Query 64 GRGGEGF--EVSKTGDPRIGLVGFPSVGKSTLLNKLT 98
G G+ F E+ P IGL+GFP+ GKSTLL L
Sbjct 146 GSKGDTFDVEIHLKLRPNIGLLGFPNAGKSTLLKALV 182
> YGR210c
Length=411
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 16/22 (72%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 77 DPRIGLVGFPSVGKSTLLNKLT 98
DP IG+VG PS GKST LN LT
Sbjct 4 DPLIGIVGKPSSGKSTTLNSLT 25
> CE26362
Length=506
Score = 33.1 bits (74), Expect = 0.14, Method: Composition-based stats.
Identities = 12/20 (60%), Positives = 18/20 (90%), Gaps = 0/20 (0%)
Query 80 IGLVGFPSVGKSTLLNKLTG 99
+G+VG+P+VGKS+ +NKL G
Sbjct 253 VGMVGYPNVGKSSTINKLAG 272
> Hs9558757
Length=396
Score = 32.7 bits (73), Expect = 0.15, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 26/41 (63%), Gaps = 5/41 (12%)
Query 65 RGGEGFE----VSKTGDP-RIGLVGFPSVGKSTLLNKLTGT 100
+GG+G + + + G +IG+VG P+VGKST N LT +
Sbjct 5 KGGDGIKPPPIIGRFGTSLKIGIVGLPNVGKSTFFNVLTNS 45
> 7298708
Length=575
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 13/20 (65%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 80 IGLVGFPSVGKSTLLNKLTG 99
IG +GFP+VGKS+L+N L G
Sbjct 335 IGCIGFPNVGKSSLINALKG 354
> ECU06g1190
Length=369
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 15/26 (57%), Positives = 20/26 (76%), Gaps = 0/26 (0%)
Query 73 SKTGDPRIGLVGFPSVGKSTLLNKLT 98
SK+ + +G+VG P+VGKSTL N LT
Sbjct 12 SKSNNLSMGIVGLPNVGKSTLFNFLT 37
> 7291084
Length=399
Score = 32.3 bits (72), Expect = 0.21, Method: Composition-based stats.
Identities = 14/20 (70%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 79 RIGLVGFPSVGKSTLLNKLT 98
RIG+VG P+VGKST N LT
Sbjct 23 RIGIVGVPNVGKSTFFNVLT 42
> Hs14149720
Length=658
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 74 KTGDPRIGLVGFPSVGKSTLLNKLTG 99
K G +GLVG+P+VGKS+ +N + G
Sbjct 383 KDGQLTVGLVGYPNVGKSSTINTIMG 408
> YGL099w
Length=640
Score = 31.6 bits (70), Expect = 0.36, Method: Composition-based stats.
Identities = 13/20 (65%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 80 IGLVGFPSVGKSTLLNKLTG 99
IGLVG+P+VGKS+ +N L G
Sbjct 339 IGLVGYPNVGKSSTINSLVG 358
> SPAC3F10.16c
Length=616
Score = 31.6 bits (70), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 12/21 (57%), Positives = 17/21 (80%), Gaps = 0/21 (0%)
Query 80 IGLVGFPSVGKSTLLNKLTGT 100
GLVG+P+VGKS+ +N L G+
Sbjct 304 FGLVGYPNVGKSSTINALVGS 324
> At4g02790
Length=375
Score = 31.6 bits (70), Expect = 0.36, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 37/65 (56%), Gaps = 9/65 (13%)
Query 33 TNFHLGLLKAKLAKLRGQLLEGSSSSSSSRSGRGGEGFEVSKTGDPRIGLVGFPSVGKST 92
TN LG+ KL +L + S + +G+ E + ++ R G++G+P+VGKS+
Sbjct 181 TNGKLGMGAMKLGRL-------AKSLAGDVNGKRREKGLLPRS--VRAGIIGYPNVGKSS 231
Query 93 LLNKL 97
L+N+L
Sbjct 232 LINRL 236
> At2g27200
Length=537
Score = 31.2 bits (69), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 11/20 (55%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 80 IGLVGFPSVGKSTLLNKLTG 99
+G VG+P+VGKS+ +N L G
Sbjct 307 VGFVGYPNVGKSSTINALVG 326
> At3g12080
Length=659
Score = 31.2 bits (69), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 12/21 (57%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 79 RIGLVGFPSVGKSTLLNKLTG 99
R+ +VG P+VGKS L N+L G
Sbjct 156 RVAIVGRPNVGKSALFNRLVG 176
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 11/21 (52%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 78 PRIGLVGFPSVGKSTLLNKLT 98
P I ++G P+VGKS++LN L
Sbjct 365 PAIAIIGRPNVGKSSILNALV 385
> SPBC428.15
Length=409
Score = 30.8 bits (68), Expect = 0.57, Method: Composition-based stats.
Identities = 15/22 (68%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 77 DPRIGLVGFPSVGKSTLLNKLT 98
D IG VG PS GKST+LN LT
Sbjct 4 DILIGFVGKPSSGKSTMLNALT 25
> Hs9506611
Length=582
Score = 30.8 bits (68), Expect = 0.61, Method: Composition-based stats.
Identities = 12/19 (63%), Positives = 17/19 (89%), Gaps = 0/19 (0%)
Query 79 RIGLVGFPSVGKSTLLNKL 97
R+G+VG P+VGKS+L+N L
Sbjct 254 RVGVVGLPNVGKSSLINSL 272
> CE12276
Length=358
Score = 30.8 bits (68), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 24/37 (64%), Gaps = 4/37 (10%)
Query 66 GGEGFEVSKTGDPRI----GLVGFPSVGKSTLLNKLT 98
GGEG E+ + R+ GLVGFP+ GKS+LL ++
Sbjct 173 GGEGEELIYDVEMRVMATAGLVGFPNAGKSSLLRAIS 209
> SPACUNK4.13c
Length=407
Score = 30.8 bits (68), Expect = 0.64, Method: Composition-based stats.
Identities = 13/22 (59%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 79 RIGLVGFPSVGKSTLLNKLTGT 100
+IG+VG P++GKSTL LT T
Sbjct 47 KIGIVGMPNIGKSTLFQILTKT 68
> SPBC26H8.08c
Length=470
Score = 30.4 bits (67), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 9/20 (45%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 80 IGLVGFPSVGKSTLLNKLTG 99
+G++G+P+VGKS+++N L
Sbjct 272 VGVIGYPNVGKSSVINALVN 291
> SPAC6F6.03c
Length=537
Score = 30.4 bits (67), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 10/18 (55%), Positives = 16/18 (88%), Gaps = 0/18 (0%)
Query 80 IGLVGFPSVGKSTLLNKL 97
+GL+GFP+ GKS+++N L
Sbjct 313 VGLIGFPNAGKSSIINTL 330
> At1g52980
Length=576
Score = 30.4 bits (67), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 10/18 (55%), Positives = 16/18 (88%), Gaps = 0/18 (0%)
Query 80 IGLVGFPSVGKSTLLNKL 97
+G VG+P+VGKS+++N L
Sbjct 312 VGFVGYPNVGKSSVINTL 329
> At5g45250
Length=1217
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 12/22 (54%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 80 IGLVGFPSVGKSTLLNKLTGTF 101
IG+VG P +GK+TLL +L T+
Sbjct 232 IGVVGMPGIGKTTLLKELYKTW 253
> CE20083
Length=698
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 10/18 (55%), Positives = 16/18 (88%), Gaps = 0/18 (0%)
Query 80 IGLVGFPSVGKSTLLNKL 97
+G +G+P+VGKS+L+N L
Sbjct 328 VGFIGYPNVGKSSLVNTL 345
> CE20084
Length=639
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 10/18 (55%), Positives = 16/18 (88%), Gaps = 0/18 (0%)
Query 80 IGLVGFPSVGKSTLLNKL 97
+G +G+P+VGKS+L+N L
Sbjct 269 VGFIGYPNVGKSSLVNTL 286
> YER006w
Length=520
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 9/18 (50%), Positives = 17/18 (94%), Gaps = 0/18 (0%)
Query 80 IGLVGFPSVGKSTLLNKL 97
+G++G+P+VGKS+++N L
Sbjct 283 VGVIGYPNVGKSSVINAL 300
Lambda K H
0.308 0.128 0.333
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184494980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40