bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2462_orf1
Length=249
Score E
Sequences producing significant alignments: (Bits) Value
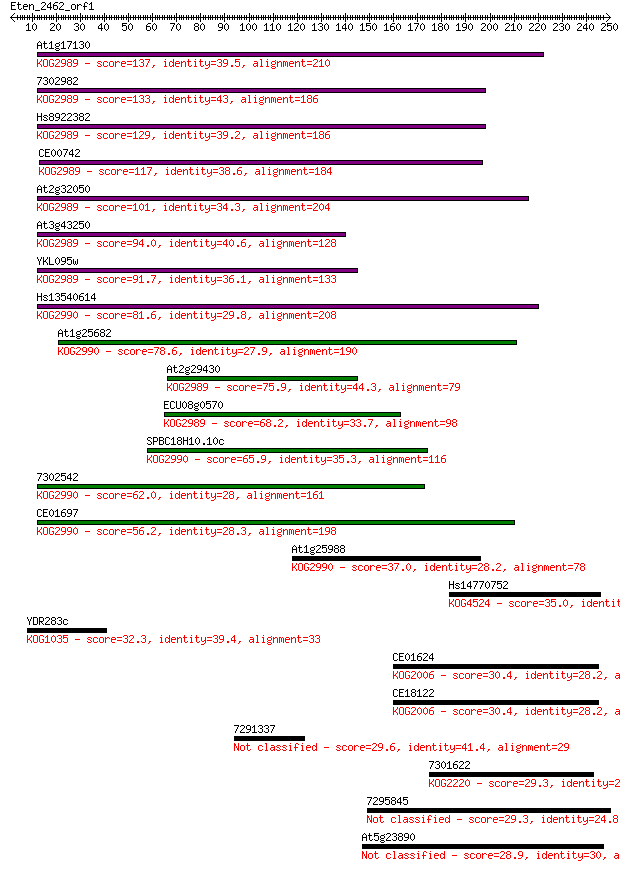
At1g17130 137 2e-32
7302982 133 4e-31
Hs8922382 129 6e-30
CE00742 117 2e-26
At2g32050 101 1e-21
At3g43250 94.0 3e-19
YKL095w 91.7 1e-18
Hs13540614 81.6 1e-15
At1g25682 78.6 1e-14
At2g29430 75.9 7e-14
ECU08g0570 68.2 1e-11
SPBC18H10.10c 65.9 8e-11
7302542 62.0 1e-09
CE01697 56.2 6e-08
At1g25988 37.0 0.035
Hs14770752 35.0 0.13
YDR283c 32.3 0.97
CE01624 30.4 3.6
CE18122 30.4 3.6
7291337 29.6 5.4
7301622 29.3 7.1
7295845 29.3 8.2
At5g23890 28.9 9.4
> At1g17130
Length=331
Score = 137 bits (345), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 83/218 (38%), Positives = 121/218 (55%), Gaps = 35/218 (16%)
Query 12 MADRKVLVKYYPPDFDFDKLQAKKKILRIQQQQLKRRRGDVFDLPQKRHKNKVMNVRMMY 71
M +RKVL KYYPPDFD KLQ R+ +R KN+ + VRMM
Sbjct 1 MGERKVLNKYYPPDFDPAKLQ------RL-----------------RRPKNQQIKVRMML 37
Query 72 PFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFKTDPQNCD 131
P +++CGTC +IY GTKFNSR E V GE YLGI +RFY +C C E+ KTDPQN D
Sbjct 38 PMSVRCGTCGNYIYKGTKFNSRKEDVIGETYLGIQIFRFYFKCTKCSAELTMKTDPQNSD 97
Query 132 YVLEWGGTRMCDPLR--DQALAAERMQKEEEEKIAMDKISQVEASRQGLQQEVLALEQLA 189
Y++E G +R +P R D+ + ++ +++ EE D + +E ++E+ + L
Sbjct 98 YIVESGASRNYEPWRAEDEEVDKDKQKRDAEE--MGDAMKSLENRTLDSKREMDIIAALD 155
Query 190 ELRRLNRRLLDRTAASDAALNFLRQ------RRIKDED 221
E++ + R T + DA L L++ +RI++ED
Sbjct 156 EMKSMKSRHA--TVSVDAMLEALQRTGAEKVKRIEEED 191
> 7302982
Length=313
Score = 133 bits (334), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 80/186 (43%), Positives = 103/186 (55%), Gaps = 23/186 (12%)
Query 12 MADRKVLVKYYPPDFDFDKLQAKKKILRIQQQQLKRRRGDVFDLPQKRHKNKVMNVRMMY 71
M++RKVL KYYPPDFD KI R+ K KN+ VR+M
Sbjct 1 MSERKVLNKYYPPDFD------PSKIPRM-----------------KLAKNRQYTVRLMA 37
Query 72 PFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFKTDPQNCD 131
PF ++C TC E+IY G KFN+R E VE E YLGI +RFY +C C EI FKTDPQN D
Sbjct 38 PFNMRCKTCGEYIYKGKKFNARKEDVENETYLGIRIYRFYIKCTRCLQEISFKTDPQNTD 97
Query 132 YVLEWGGTRMCDPLRDQALAAERMQKEEEEKIAMDKISQVEASRQGLQQEVLALEQLAEL 191
Y +E G TR L+ A R ++E ++ A + + +E Q + E+ +E L EL
Sbjct 98 YEIEAGATRNFMALKLAEEQARREEQELRDEEANNPMKLLENRTQQSRNEIETIESLEEL 157
Query 192 RRLNRR 197
R LNRR
Sbjct 158 RDLNRR 163
> Hs8922382
Length=323
Score = 129 bits (324), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 73/186 (39%), Positives = 101/186 (54%), Gaps = 23/186 (12%)
Query 12 MADRKVLVKYYPPDFDFDKLQAKKKILRIQQQQLKRRRGDVFDLPQKRHKNKVMNVRMMY 71
M++RKVL KYYPPDFD K+ K LP+ R VR+M
Sbjct 1 MSERKVLNKYYPPDFDPSKIPKLK-------------------LPKDRQ----YVVRLMA 37
Query 72 PFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFKTDPQNCD 131
PF ++C TC E+IY G KFN+R E V+ E YLG+ +RFY +C C EI FKTDP+N D
Sbjct 38 PFNMRCKTCGEYIYKGKKFNARKETVQNEVYLGLPIFRFYIKCTRCLAEITFKTDPENTD 97
Query 132 YVLEWGGTRMCDPLRDQALAAERMQKEEEEKIAMDKISQVEASRQGLQQEVLALEQLAEL 191
Y +E G R + +R+QKE E++ + + +E + + E+ LE L EL
Sbjct 98 YTMEHGAARNFQAEKLLEEEEKRVQKEREDEELNNPMKVLENRTKDSKLEMEVLENLQEL 157
Query 192 RRLNRR 197
+ LN+R
Sbjct 158 KDLNQR 163
> CE00742
Length=323
Score = 117 bits (293), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 71/187 (37%), Positives = 99/187 (52%), Gaps = 30/187 (16%)
Query 13 ADRKVLVKYYPPDFDFDKLQAKKKILRIQQQQLKRRRGDVFDLPQKRHKNKVMNVRMMYP 72
+RKV KYYPPDFD KI R + Q KN+ R+M P
Sbjct 4 TERKVFQKYYPPDFD------PSKIPRAKGQ-----------------KNRQFVQRVMTP 40
Query 73 FTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFKTDPQNCDY 132
F ++C TC E+IY G KFN + E EGE YLG+ +RFY RCP+C EI FKTD +NCDY
Sbjct 41 FNMQCNTCHEYIYKGKKFNMKRETAEGETYLGLKLFRFYFRCPNCLAEITFKTDLENCDY 100
Query 133 VLEWGGTRMCDPLR---DQALAAERMQKEEEEKIAMDKISQVEASRQGLQQEVLALEQLA 189
E G TR+ + ++ DQ ++ E+E + A D + +E +QE+ + L
Sbjct 101 QNEHGATRLFEAVKLYQDQ----QKQVDEQEAEDAKDPMKMLEKRTMQSRQEMEEMGNLE 156
Query 190 ELRRLNR 196
+L+ +R
Sbjct 157 DLQESSR 163
> At2g32050
Length=254
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 70/206 (33%), Positives = 100/206 (48%), Gaps = 39/206 (18%)
Query 12 MADRKVLVKYYPPDFDFDKLQAKKKILRIQQQQLKRRRGDVFDLPQKRHKNKVMNVRMMY 71
M +RK L KYYPP+FD K+I RI ++ KN+ +R M
Sbjct 1 MGERKGLNKYYPPNFD------PKQIPRI-----------------RKPKNQQRKIRSMV 37
Query 72 PFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFKTDPQNCD 131
P ++C TC ++ GTK N R E V GE YLGI RFY +C C E+ KTDP+N
Sbjct 38 PLRIRCNTCGNYMSEGTKINCREENVIGETYLGIKIHRFYFKCSKCCTELILKTDPKNSS 97
Query 132 YVLEWGGTRMCDPLRDQALAAERMQKEEEEKI--AMDKISQVEASRQGLQQEVLALEQLA 189
YV E G T + D Q EEEE+ D++S +E ++EV + L
Sbjct 98 YVAESGATCVYD------------QHEEEEQAEDGGDRMSSLEKRTLVSKREVDVMAALD 145
Query 190 ELRRLNRRLLDRTAASDAALNFLRQR 215
E++ + R + + + D+ L L +R
Sbjct 146 EMKSMKSRRV--SVSVDSMLEDLSKR 169
> At3g43250
Length=249
Score = 94.0 bits (232), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 52/128 (40%), Positives = 66/128 (51%), Gaps = 23/128 (17%)
Query 12 MADRKVLVKYYPPDFDFDKLQAKKKILRIQQQQLKRRRGDVFDLPQKRHKNKVMNVRMMY 71
M +RK L KYYPPDFD KKI RI K+ KN+ +R M
Sbjct 1 MGERKNLNKYYPPDFD------PKKIHRI-----------------KKPKNQQKKIRFML 37
Query 72 PFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFKTDPQNCD 131
P ++C TC ++ GTKFN R E V E YLG+ RFY +C C E+ KTDP+N
Sbjct 38 PVRVRCNTCGNYMSEGTKFNCRQEDVITETYLGLKIHRFYIKCTKCLAELTIKTDPKNHS 97
Query 132 YVLEWGGT 139
Y +E G +
Sbjct 98 YTVESGAS 105
> YKL095w
Length=278
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 48/134 (35%), Positives = 72/134 (53%), Gaps = 17/134 (12%)
Query 12 MADRKVLVKYYPPDFDFDKLQAKKKILRIQQQQLKRRRGDVFDLPQKRHKNKVMNVRMMY 71
M++RK + KYYPPD++ ++ ++L R+ K H ++R+M
Sbjct 1 MSERKAINKYYPPDYN-----------PLEAEKLSRKMAKKLKTMNKSH----ASIRLMT 45
Query 72 PFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIK-WRFYGRCPHCRGEICFKTDPQNC 130
PF+++C C+E+I KFN + E ++ E YL IK +R CP C I F+TDP N
Sbjct 46 PFSMRCLECNEYIPKSRKFNGKKELLK-EKYLDSIKIYRLTISCPRCANSIAFRTDPGNS 104
Query 131 DYVLEWGGTRMCDP 144
DYV+E GG R P
Sbjct 105 DYVMEVGGVRNYVP 118
> Hs13540614
Length=396
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 62/214 (28%), Positives = 100/214 (46%), Gaps = 28/214 (13%)
Query 12 MADRKVLVKYYPPDFDFDKLQAKKKILRIQQQQLKRRRGDVFDLPQKRHKNKVMNVRMMY 71
M +RK + KYYPPDF+ +K + + R R R ++ ++ +R
Sbjct 1 MGERKGVNKYYPPDFNPEKHGS---LNRYHNSHPLRERA-------RKLSQGILIIRFEM 50
Query 72 PFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFKTDPQNCD 131
P+ + C C I +G ++N+ +KV G Y I +RF +C C I +TDP NCD
Sbjct 51 PYNIWCDGCKNHIGMGVRYNAEKKKV-GNYYTTPI-YRFRMKCHLCVNYIEMQTDPANCD 108
Query 132 YVLEWGGTR------MCDPLRDQALAAERMQKEEEEKIAMDKISQVEASRQGLQQEVLAL 185
YV+ G R M D +Q L E +K++ E AM ++ EA R L++ + L
Sbjct 109 YVIVSGAQRKEERWDMAD--NEQVLTTEHEKKQKLETDAMFRLEHGEADRSTLKKALPTL 166
Query 186 EQLAELRRLNRRLLDRTAASDAALNFLRQRRIKD 219
+ E + + D ALN + +RR ++
Sbjct 167 SHIQEAQSAWK--------DDFALNSMLRRRFRE 192
> At1g25682
Length=310
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 53/190 (27%), Positives = 87/190 (45%), Gaps = 17/190 (8%)
Query 21 YYPPDFDFDKLQAKKKILRIQQQQLKRRRGDVFDLPQKRHKNKVMNVRMMYPFTLKCGTC 80
YYPP++ D+ + + Q Q R R K+ ++ +R P+ + CG C
Sbjct 13 YYPPEWTPDQ----GSLNKFQGQHPLRERA-------KKIGEGILVIRFEMPYNIWCGGC 61
Query 81 SEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFKTDPQNCDYVLEWGGTR 140
S I G +FN+ EK + +Y W F + P C+ EI +TDPQNC+YV+ G +
Sbjct 62 SSMIAKGVRFNA--EKKQVGNYYSTKIWSFAMKSPCCKHEIVIQTDPQNCEYVITSGAQK 119
Query 141 MCDPLRDQALAAERMQKEEEEKIAMDKISQVEASRQGLQQEVLALEQLAELRRLNRRLLD 200
+ + + E+E+ D ++E LQ++ A L L+R++ D
Sbjct 120 KVEEYEAEDAETMELTAEQEKGKLADPFYRLEHQEVDLQKKKAAEPLLVRLQRVS----D 175
Query 201 RTAASDAALN 210
A D +LN
Sbjct 176 ARHADDYSLN 185
> At2g29430
Length=84
Score = 75.9 bits (185), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 48/79 (60%), Gaps = 0/79 (0%)
Query 66 NVRMMYPFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFKT 125
N + P L+C C + GTKF SRVE V GE YLGI +RF +C + E+ F+T
Sbjct 5 NADLSLPMRLQCNNCDNIMSKGTKFTSRVEDVIGETYLGIKIFRFQIQCTNGSHEMKFRT 64
Query 126 DPQNCDYVLEWGGTRMCDP 144
DP+N D+++E G TR+ P
Sbjct 65 DPKNADFIIESGATRLLLP 83
> ECU08g0570
Length=162
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 33/98 (33%), Positives = 54/98 (55%), Gaps = 10/98 (10%)
Query 65 MNVRMMYPFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFK 124
++ R+ PFT++C +C +IY + N+ E + +DYLG+ +RF+ C C +C +
Sbjct 34 LSTRLATPFTIRCLSCGSYIYKNKRHNAVREIAQDKDYLGVESYRFHIICTGCGRALCIR 93
Query 125 TDPQNCDYVLEWGGTRMCDPLRDQALAAERMQKEEEEK 162
TDP+N YV E G R+ E + K++EEK
Sbjct 94 TDPKNGVYVTESGCIRI----------DEEVTKQDEEK 121
> SPBC18H10.10c
Length=294
Score = 65.9 bits (159), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 41/117 (35%), Positives = 58/117 (49%), Gaps = 6/117 (5%)
Query 58 KRHKNKVMNV-RMMYPFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPH 116
KR +K+ NV R PF + C C I GT+FN+ V+K G Y I W F +C
Sbjct 13 KRKFDKLRNVIRFEMPFPVWCNNCENIIQQGTRFNA-VKKEIGSYYTTKI-WSFSLKCHL 70
Query 117 CRGEICFKTDPQNCDYVLEWGGTRMCDPLRDQALAAERMQKEEEEKIAMDKISQVEA 173
C I TDP+N +Y++ GG R +P Q + + E +EK+ D I +E
Sbjct 71 CSNPIDVHTDPKNTEYIVASGGRRKIEP---QDINERPAKAENDEKVPSDAIEALET 124
> 7302542
Length=316
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 45/164 (27%), Positives = 75/164 (45%), Gaps = 18/164 (10%)
Query 12 MADRKVLVKYYPPDFDFDKLQAKKKILRIQQQQLKRRRGDVFDLPQKRHKNKVMNVRMMY 71
M +RK KYYPPD+D K + + Q R R L ++ +R
Sbjct 1 MGERKGQNKYYPPDYD----PKKGGLNKFQGTHALRERARKIHL-------GIIIIRFEM 49
Query 72 PFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFKTDPQNCD 131
P+ + C C I +G ++N+ EK + Y ++F +C C +TDP N D
Sbjct 50 PYNIWCDGCKNHIGMGVRYNA--EKTKVGMYYTTPVFKFRMKCHLCDNHFEIQTDPGNLD 107
Query 132 YVLEWGGTRM---CDPLRDQALAAERMQKEEEEKIAMDKISQVE 172
YV+ G R DPL+++ + E KE ++++ D + ++E
Sbjct 108 YVILSGARRQENRWDPLQNEQVVPE--TKEVQKRLFDDAMYKLE 149
> CE01697
Length=369
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 56/219 (25%), Positives = 89/219 (40%), Gaps = 33/219 (15%)
Query 12 MADRKVLVKYYPPDFDFDKLQAKKKILRIQQQQLKRRRGDVFDLPQKRHKNKVMNVRMMY 71
M +RK YYPPDF++ + K + R R D ++ +R
Sbjct 1 MGERKGQNFYYPPDFNY---KTHKSLNGYHGTHALRERAKKID-------QGILVIRFEM 50
Query 72 PFTLKCGTCSEFIYVGTKFNSRVEKVEGEDYLGIIKWRFYGRCPHCRGEICFKTDPQNCD 131
PF + C C + +G ++N+ +K+ Y F +C C +TDP+N D
Sbjct 51 PFNIWCLGCHNHVGMGVRYNAEKKKIGM--YYTTPLHEFRMKCHLCDNYYVIRTDPKNFD 108
Query 132 YVLEWGGTRM---CDPLR-----------DQALAAERMQKEEEEKIAMDK-------ISQ 170
Y L G +R DP Q LAA+ M K+E E DK + +
Sbjct 109 YELVEGCSRQELRFDPTDIAQIGAVDRGFTQKLAADAMFKKEHEAEDKDKAATEEGRVDK 168
Query 171 VEASRQGLQQEVLALEQLAELRRLNRRLLDRTAASDAAL 209
+E ++ ++ + A L R ++ L+ T A DA L
Sbjct 169 LEWIQERMRDDFTANSFLRAQFRNEKKSLNETRARDANL 207
> At1g25988
Length=180
Score = 37.0 bits (84), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 37/78 (47%), Gaps = 0/78 (0%)
Query 118 RGEICFKTDPQNCDYVLEWGGTRMCDPLRDQALAAERMQKEEEEKIAMDKISQVEASRQG 177
RG KTDPQNC+YV+ G + + + + E+E+ D ++E
Sbjct 21 RGNFGHKTDPQNCEYVITSGAQKKVEEYEAEDAETMELTAEQEKGKLADPFYRLEHQEVD 80
Query 178 LQQEVLALEQLAELRRLN 195
LQ++ A L L+R++
Sbjct 81 LQKKKAAEPFLVRLQRVS 98
> Hs14770752
Length=1089
Score = 35.0 bits (79), Expect = 0.13, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 31/67 (46%), Gaps = 4/67 (5%)
Query 183 LALEQLAELRRLNRRLLDRTAASDAALNFLRQRRIKDEDMEGSP----AQEWTEIEKDEL 238
L +A L+RL++ L+ A + + +RR +D+ SP Q W I++
Sbjct 417 FVLNSVAHLQRLSKALIQVLELDVADIKIVEERRWNSDDLNASPKTSATQPWNRIQRRYF 476
Query 239 KFFRAAR 245
+FF R
Sbjct 477 RFFTDER 483
> YDR283c
Length=1659
Score = 32.3 bits (72), Expect = 0.97, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 8 RVNKMADRKVLVKYYPPDFDFDKLQAKKKILRI 40
RVN + + + +PPDFD +K++ +KKI+R+
Sbjct 928 RVNILKKLRSVSIEFPPDFDDNKMKVEKKIIRL 960
> CE01624
Length=1941
Score = 30.4 bits (67), Expect = 3.6, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 43/85 (50%), Gaps = 11/85 (12%)
Query 160 EEKIAMDKISQVEASRQGLQQEVLALEQLAELRRLNRRLLDRTAASDAALNFLRQRRIKD 219
E+K+ ++SQ AS GL + L + + R+L R ++D A + RI++
Sbjct 1626 EKKLGNIRVSQSTASLNGLLNDSLDNIKKPKARKLERTVVDLNGA---------RSRIRE 1676
Query 220 EDMEGSPAQEWTEIEKDELKFFRAA 244
D+E S +E EI++ L+ R +
Sbjct 1677 TDIEAS--EEDVEIQEAVLERVRGS 1699
> CE18122
Length=1943
Score = 30.4 bits (67), Expect = 3.6, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 43/85 (50%), Gaps = 11/85 (12%)
Query 160 EEKIAMDKISQVEASRQGLQQEVLALEQLAELRRLNRRLLDRTAASDAALNFLRQRRIKD 219
E+K+ ++SQ AS GL + L + + R+L R ++D A + RI++
Sbjct 1628 EKKLGNIRVSQSTASLNGLLNDSLDNIKKPKARKLERTVVDLNGA---------RSRIRE 1678
Query 220 EDMEGSPAQEWTEIEKDELKFFRAA 244
D+E S +E EI++ L+ R +
Sbjct 1679 TDIEAS--EEDVEIQEAVLERVRGS 1701
> 7291337
Length=691
Score = 29.6 bits (65), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 94 VEKVEGEDYLGIIKWRFYGRCPHCRGEIC 122
+ +EGEDYL +K +C C GE+C
Sbjct 314 LTSLEGEDYLDCLKDSNITKCSRCTGELC 342
> 7301622
Length=836
Score = 29.3 bits (64), Expect = 7.1, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 29/69 (42%), Gaps = 7/69 (10%)
Query 175 RQGLQQEVLALEQLAELRRLNRRLLDRTAASDAALNFLRQRRIKDEDMEGSPAQEWTEIE 234
+ G++ L+ L EL NR +LD T L + R D + WT I
Sbjct 430 KGGIENVQTMLKDLPELLNRNREILDETE------RLLDEERDSDNQLRAQFKDRWTRIS 483
Query 235 KDEL-KFFR 242
D+L + FR
Sbjct 484 SDKLTEMFR 492
> 7295845
Length=2168
Score = 29.3 bits (64), Expect = 8.2, Method: Composition-based stats.
Identities = 25/107 (23%), Positives = 51/107 (47%), Gaps = 10/107 (9%)
Query 149 ALAAERMQKEEEEKIAMD-KISQVEASRQGLQQEVLALE-----QLAELRRLNRRLLDRT 202
AL E ++ E++ + ++ QVE QG ++E++ E +LAE + + L+DR
Sbjct 1648 ALKQEHLRTAEQKIQELQTRLQQVETEEQGHREELIRKENIHTARLAEANQREQDLIDRV 1707
Query 203 AASDAALNFLRQRRIKDEDMEGSPAQEWTEIEKDELKFFRAARELDS 249
+ LN L+ + +E ++ + +DE+ R + + S
Sbjct 1708 KSLTKELNTLKANKEHNE----RDLRDRLALSQDEISVLRTSSQRRS 1750
> At5g23890
Length=946
Score = 28.9 bits (63), Expect = 9.4, Method: Composition-based stats.
Identities = 30/117 (25%), Positives = 52/117 (44%), Gaps = 17/117 (14%)
Query 147 DQALAAERMQKEEEEKIAMDKISQVEASRQGLQQEVLAL-----------EQLAELRRLN 195
++ L+ ER + E EK+A ++E R+ ++E LAL E L+ LRR
Sbjct 672 EKELSMEREKIEAVEKMAELAKVELEQLREKREEENLALVKERAAVESEMEVLSRLRRDA 731
Query 196 RRLLDRTAASDAALNFLRQR------RIKDEDMEGSPAQEWTEIEKDELKFFRAARE 246
L+ ++ A + F ++R ++E S Q E+E+ L R+ E
Sbjct 732 EEKLEDLMSNKAEITFEKERVFNLRKEAEEESQRISKLQYELEVERKALSMARSWAE 788
Lambda K H
0.322 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5080928138
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40