bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2441_orf1
Length=326
Score E
Sequences producing significant alignments: (Bits) Value
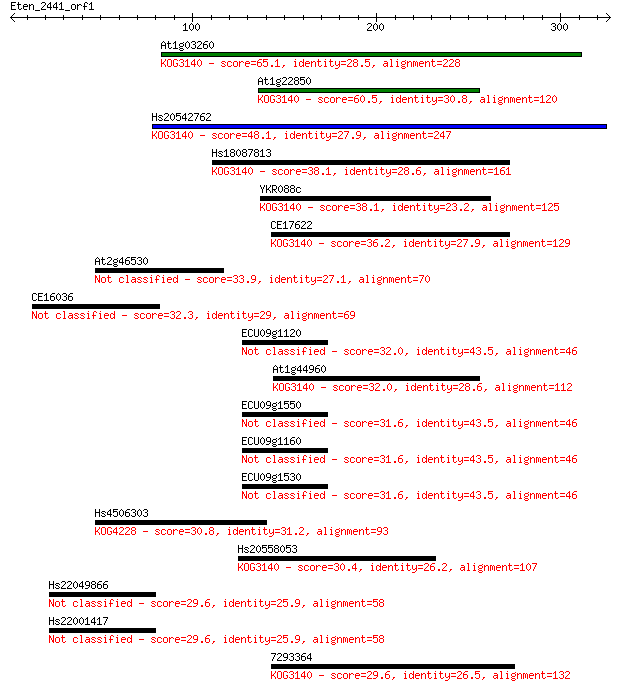
At1g03260 65.1 2e-10
At1g22850 60.5 4e-09
Hs20542762 48.1 2e-05
Hs18087813 38.1 0.023
YKR088c 38.1 0.025
CE17622 36.2 0.094
At2g46530 33.9 0.46
CE16036 32.3 1.5
ECU09g1120 32.0 1.8
At1g44960 32.0 2.0
ECU09g1550 31.6 2.2
ECU09g1160 31.6 2.2
ECU09g1530 31.6 2.2
Hs4506303 30.8 3.8
Hs20558053 30.4 5.9
Hs22049866 29.6 9.1
Hs22001417 29.6 9.1
7293364 29.6 9.7
> At1g03260
Length=269
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 65/235 (27%), Positives = 115/235 (48%), Gaps = 24/235 (10%)
Query 83 PTCGRVQLGLLGVAVSACFIILFLLPFKDVLRQVVEHQKG-SPLTYALTYVVAGLV-VPA 140
P+ R+ + LL + V+ ++FL KD L + E PL AL Y+ +V VPA
Sbjct 5 PSTFRIAISLL-LLVAIVSAVIFLPKLKDFLLWIKEDLGPFGPLALALAYIPLTIVAVPA 63
Query 141 PLLSVLAGVLMGPSLLAVFVIMCGSMGAACLAFFISRFMLRRFVVNRFVRRSKQLQAIDL 200
+L++ G L G + V + ++GA AF + R + + +V ++ ++ + QA+ +
Sbjct 64 SVLTLGGGYLFGLPVGFVADSLGATLGATA-AFLLGRTIGKSYVTSK-IKHYPKFQAVSV 121
Query 201 ALQKDSVKLVLCTRM--ILPFTFNNYFLGTTPISAATFAVST---LVTGIPFAVLYAIIG 255
A+QK K+VL R+ ILPF NY L TP+ + ++T ++ I FA++Y +G
Sbjct 122 AIQKSGFKIVLLLRVVPILPFNMLNYLLSVTPVRLGEYMLATWLGMMQPITFALVY--VG 179
Query 256 GQLQSLDNALAAKSFELQGADVDVFGFFTMSKRHLEVIGICGGILLCFFVVRTIK 310
L+ L + +V VF + M ++G+ ++L + R K
Sbjct 180 TTLKDLSDITHGWH------EVSVFRWVIM------MVGVALAVILIICITRVAK 222
> At1g22850
Length=344
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 68/122 (55%), Gaps = 4/122 (3%)
Query 136 LVVPAPLLSVLAGVLMGPSLLAVFVIMCGSMGAACLAFFISRFMLRRFVVNRFVRRSKQL 195
L +PA L++ AG+L GP + + V + G+M AA +AF I+R+ R ++ + V +K+
Sbjct 166 LAIPALPLTMSAGLLFGPLIGTIIVSISGTM-AASVAFLIARYFARERIL-KLVEDNKKF 223
Query 196 QAIDLALQKDSVKLVLCTRM--ILPFTFNNYFLGTTPISAATFAVSTLVTGIPFAVLYAI 253
AID A+ ++ ++V R+ +LPF+ NY G T + + + + + +P + Y
Sbjct 224 LAIDKAIGENGFRVVTLLRLSPLLPFSLGNYLYGLTSVKFVPYVLGSWLGMLPGSWAYVS 283
Query 254 IG 255
G
Sbjct 284 AG 285
> Hs20542762
Length=343
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 69/260 (26%), Positives = 120/260 (46%), Gaps = 41/260 (15%)
Query 78 PDELPPTCGRVQLGLLGVAVSACFIILFLLPFKDVLRQVVEH-----QKGSPLTYALTYV 132
P E PTC L L+ V + CF L L +R+ + H + L L +V
Sbjct 74 PPEWAPTCWCRSLVLVCVLAALCFASLAL------VRRYLHHLLLWVESLDSLLGVLLFV 127
Query 133 VAGLVVPAP------LLSVLAGVLMGPSLLAVFVIMCGSMGAACLAFFISRFMLRRFVVN 186
V +VV P +L+V AG L G +L + ++M G + +A + + +L +V
Sbjct 128 VGFIVVSFPCGWGYIVLNVAAGYLYG-FVLGMGLMMVGVLIGTFIAHVVCKRLLTAWVAA 186
Query 187 RFVRRSKQLQAIDLALQKDS-VKLVLCTRMI-LPFTFNNYFLGTTPISAATFAVSTLVTG 244
R ++ S++L A+ ++ S +K+V R+ +PF N T +S + +++ V
Sbjct 187 R-IQSSEKLSAVIRVVEGGSGLKVVALARLTPIPFGLQNAVFSITDLSLPNYLMASSVGL 245
Query 245 IPFAVLYAIIGGQLQSLDNALAAKSFELQGADVDVFGFFTMSKRHLEVIGICGGILLCFF 304
+P +L + +G L+++++ +A +S V G+F + + IG L F+
Sbjct 246 LPTQLLNSYLGTTLRTMEDVIAEQS---------VSGYFVFCLQIIISIG------LMFY 290
Query 305 VVRTIKRFAAQVVLEAQEVA 324
VV AQV L A VA
Sbjct 291 VVHR-----AQVELNAAIVA 305
> Hs18087813
Length=264
Score = 38.1 bits (87), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 46/167 (27%), Positives = 76/167 (45%), Gaps = 8/167 (4%)
Query 111 DVLRQV-VEHQKGSPLTYALTYVVA-GLVVP-APLLSVLAGVLMGPSLLAVFVIMCGSMG 167
+VLR+ EHQ L + Y+ G +P + L+VLAG L GP L + + S+G
Sbjct 56 EVLREYRKEHQAYVFLLFCGAYLYKQGFAIPGSSFLNVLAGALFGPWLGLLLCCVLTSVG 115
Query 168 AACLAFFISRFMLRRFVVNRFVRRSKQLQAIDLALQKDSVKLVLCTRMILPFTFNNYFLG 227
A C + +S ++ VV+ F + LQ + ++S+ L + P T N +
Sbjct 116 ATC-CYLLSSIFGKQLVVSYFPDKVALLQR-KVEENRNSLFFFLLFLRLFPMTPNWFLNL 173
Query 228 TTP---ISAATFAVSTLVTGIPFAVLYAIIGGQLQSLDNALAAKSFE 271
+ P I F S L+ IP+ + G L +L + A S++
Sbjct 174 SAPILNIPIVQFFFSVLIGLIPYNFICVQTGSILSTLTSLDALFSWD 220
> YKR088c
Length=337
Score = 38.1 bits (87), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 29/128 (22%), Positives = 61/128 (47%), Gaps = 5/128 (3%)
Query 137 VVPAPLLSVLAGVLMGPSLLAVFVIMCGSMGAACLAFFISRFMLRRFVVNRFVRRSKQLQ 196
++ LLS G++ G S + GS+ + +F + + +L + V +++ +
Sbjct 152 MIGYSLLSTTTGLIYGVSFEGWVTLALGSVTGSIASFVVFKTILHS-RAEKLVHLNRRFE 210
Query 197 AIDLALQKDSVKLVLCTRMILPFTF---NNYFLGTTPISAATFAVSTLVTGIPFAVLYAI 253
A+ LQ+++ +L + PF + N G IS F+++ ++T P +Y
Sbjct 211 ALASILQENNSYWILALLRLCPFPYSLTNGAIAGVYGISVRNFSIANIIT-TPKLFIYLF 269
Query 254 IGGQLQSL 261
IG +++SL
Sbjct 270 IGSRVKSL 277
> CE17622
Length=246
Score = 36.2 bits (82), Expect = 0.094, Method: Compositional matrix adjust.
Identities = 36/132 (27%), Positives = 58/132 (43%), Gaps = 5/132 (3%)
Query 143 LSVLAGVLMGPSLLAVFVIMCGSMGAACLAFFISRFMLRRFVVNRFVRRSKQLQAIDLAL 202
L++L+G L + V V C + GAA + + IS+ R FV+ +F R + Q DL+
Sbjct 92 LTILSGYLFPFYVAIVLVCSCSATGAA-ICYTISKLFGRSFVLQKFPERIAKWQD-DLSK 149
Query 203 QKD---SVKLVLCTRMILPFTFNNYFLGTTPISAATFAVSTLVTGIPFAVLYAIIGGQLQ 259
+D + + L I+P N + A F T + P + LY G L+
Sbjct 150 HRDDFLNYMIFLRVTPIVPNWLINIASPVLDVPLAPFFWGTFLGVAPPSFLYIQAGSTLE 209
Query 260 SLDNALAAKSFE 271
L + A S+
Sbjct 210 QLSHTSVAWSWS 221
> At2g46530
Length=622
Score = 33.9 bits (76), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 40/74 (54%), Gaps = 4/74 (5%)
Query 47 AAVSPRERELLTV-TNLSQSFPASSGPNSSDDPDELP---PTCGRVQLGLLGVAVSACFI 102
+A P +++L++V +N+S S PNSS+ P E T R+++ + G AV
Sbjct 469 SATIPHDKQLISVDSNISDSTTKCQDPNSSNSPKEQKQQTSTRSRIKVQMQGTAVGRAVD 528
Query 103 ILFLLPFKDVLRQV 116
+ L + ++++++
Sbjct 529 LTLLRSYDELIKEL 542
> CE16036
Length=316
Score = 32.3 bits (72), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 32/70 (45%), Gaps = 1/70 (1%)
Query 13 FLPLTICAIEPLMAIFLRLFSQRHSSRRSSLEDVAAVSPRERELLTVTN-LSQSFPASSG 71
FLP T + ++ +FL+L ++H R SLE+ ++ LL TN + F
Sbjct 196 FLPWTAVLLNLIVFVFLKLRKKKHQMNRKSLENSLFINSFVVSLLLTTNYIYNHFYLMFE 255
Query 72 PNSSDDPDEL 81
P S E+
Sbjct 256 PTLSQQSPEI 265
> ECU09g1120
Length=574
Score = 32.0 bits (71), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 32/48 (66%), Gaps = 6/48 (12%)
Query 127 YALTYVVAGLVVPAPLLSVLAGVLMGPSLLAVFVIMCGS--MGAACLA 172
Y++T VAG + A +++V GVL G SL+ V V++CG+ +GA C+
Sbjct 453 YSMT--VAG--IGAVMMAVQGGVLCGMSLMQVCVVVCGNLVLGALCVG 496
> At1g44960
Length=280
Score = 32.0 bits (71), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 52/115 (45%), Gaps = 8/115 (6%)
Query 144 SVLAGVLMGPSLLAVFVIMCGSMGAACLAFFISRFMLRRFV-VNRFVRRSKQLQAIDLAL 202
S+L G L P+LL VF + AA +F+I RF+ + + +K + +
Sbjct 74 SMLFGFL--PALLCVF---SAKVLAASFSFWIGRFVFKSSTRATGWAHSNKYFNILSRGV 128
Query 203 QKDSVKLVLCTRMI-LPFTFNNYFLGTTPIS-AATFAVSTLVTGIPFAVLYAIIG 255
++D K VL R +P NY L T + A F T++ +P + A +G
Sbjct 129 ERDGWKFVLLARFSPIPSYVINYALAATEVRFVADFLFPTVIGCLPMILQNASVG 183
> ECU09g1550
Length=591
Score = 31.6 bits (70), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 32/48 (66%), Gaps = 6/48 (12%)
Query 127 YALTYVVAGLVVPAPLLSVLAGVLMGPSLLAVFVIMCGS--MGAACLA 172
Y++T VAG + A +++V GVL G SL+ V V++CG+ +GA C+
Sbjct 467 YSMT--VAG--IGAVMMAVQGGVLCGMSLMQVCVVVCGNLVLGALCVG 510
> ECU09g1160
Length=534
Score = 31.6 bits (70), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 32/48 (66%), Gaps = 6/48 (12%)
Query 127 YALTYVVAGLVVPAPLLSVLAGVLMGPSLLAVFVIMCGS--MGAACLA 172
Y++T VAG + A +++V GVL G SL+ V V++CG+ +GA C+
Sbjct 413 YSMT--VAG--IGAVMMAVQGGVLCGMSLMQVCVVVCGNLVLGALCVG 456
> ECU09g1530
Length=570
Score = 31.6 bits (70), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 32/48 (66%), Gaps = 6/48 (12%)
Query 127 YALTYVVAGLVVPAPLLSVLAGVLMGPSLLAVFVIMCGS--MGAACLA 172
Y++T VAG + A +++V GVL G SL+ V V++CG+ +GA C+
Sbjct 423 YSMT--VAG--IGAVMMAVQGGVLCGMSLMQVCVVVCGNLVLGALCVG 466
> Hs4506303
Length=802
Score = 30.8 bits (68), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 37/93 (39%), Gaps = 2/93 (2%)
Query 47 AAVSPRERELLTVTNLSQSFPASSGPNSSDDPDELPPTCGRVQLGLLGVAVSACFIILFL 106
A P E T ++FP S +S D DE P V L L V V I+L++
Sbjct 116 ARTEPWEGNSSTAATTPETFPPSGNSDSKDRRDETPIIAVMVALSSLLVIV-FIIIVLYM 174
Query 107 LPFKDVLRQVVEHQKGSPLTYALTYVVAGLVVP 139
L FK +Q H L+ T V VP
Sbjct 175 LRFKK-YKQAGSHSNSFRLSNGRTEDVEPQSVP 206
> Hs20558053
Length=291
Score = 30.4 bits (67), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 28/113 (24%), Positives = 62/113 (54%), Gaps = 12/113 (10%)
Query 125 LTYALTYV-VAGLVVPAPL-LSVLAGVLMGPSLLAVFVI-MCGSMGAA---CLAFFISRF 178
+ Y TY+ + +P + LS+L+G L P LA+F++ +C +GA+ L++ + R
Sbjct 115 VAYFATYIFLQTFAIPGSIFLSILSGFLY-PFPLALFLVCLCSGLGASFCYMLSYLVGRP 173
Query 179 MLRRFVVNRFVRRSKQLQAIDLALQKDSVKLVLCTRMILPFTFNNYFLGTTPI 231
++ +++ + V+ S+Q++ ++ + ++ R I PF N + T+P+
Sbjct 174 VVYKYLTEKAVKWSQQVE----RHREHLINYIIFLR-ITPFLPNWFINITSPV 221
> Hs22049866
Length=1106
Score = 29.6 bits (65), Expect = 9.1, Method: Composition-based stats.
Identities = 15/58 (25%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 22 EPLMAIFLRLFSQRHSSRRSSLEDVAAVSPRERELLTVTNLSQSFPASSGPNSSDDPD 79
E +++ F LFS++H+S ++S VA V E++ SQ +++ ++P+
Sbjct 948 ERMLSTFKELFSEKHASLQNSQRTVAEVQETLAEMIRQHQKSQLCKSTANGPDKNEPE 1005
> Hs22001417
Length=1508
Score = 29.6 bits (65), Expect = 9.1, Method: Composition-based stats.
Identities = 15/58 (25%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 22 EPLMAIFLRLFSQRHSSRRSSLEDVAAVSPRERELLTVTNLSQSFPASSGPNSSDDPD 79
E +++ F LFS++H+S ++S VA V E++ SQ +++ ++P+
Sbjct 1350 ERMLSTFKELFSEKHASLQNSQRTVAEVQETLAEMIRQHQKSQLCKSTANGPDKNEPE 1407
> 7293364
Length=304
Score = 29.6 bits (65), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 35/138 (25%), Positives = 58/138 (42%), Gaps = 11/138 (7%)
Query 143 LSVLAGVLMGPSLLAVFVIMCGSMGAA---CLAFFISRFMLRRFVVNRFVRRSKQLQAID 199
LS+L G L + + C ++GA L+ + R ++R F + SK ++
Sbjct 147 LSILLGFLYKFPIALFLICFCSALGATLCYTLSNLVGRRLIRHFWPKKTSEWSKHVEEY- 205
Query 200 LALQKDSV-KLVLCTRM--ILPFTFNNYFLGTTPISAATFAVSTLVTGIPFAVLYAIIGG 256
+DS+ +L RM ILP F N + FA+ T P +V+ G
Sbjct 206 ----RDSLFNYMLFLRMTPILPNWFINLASPVIGVPLHIFALGTFCGVAPPSVIAIQAGK 261
Query 257 QLQSLDNALAAKSFELQG 274
LQ + ++ A S+ G
Sbjct 262 TLQKMTSSSEAFSWTSMG 279
Lambda K H
0.330 0.141 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7636708702
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40