bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2426_orf1
Length=165
Score E
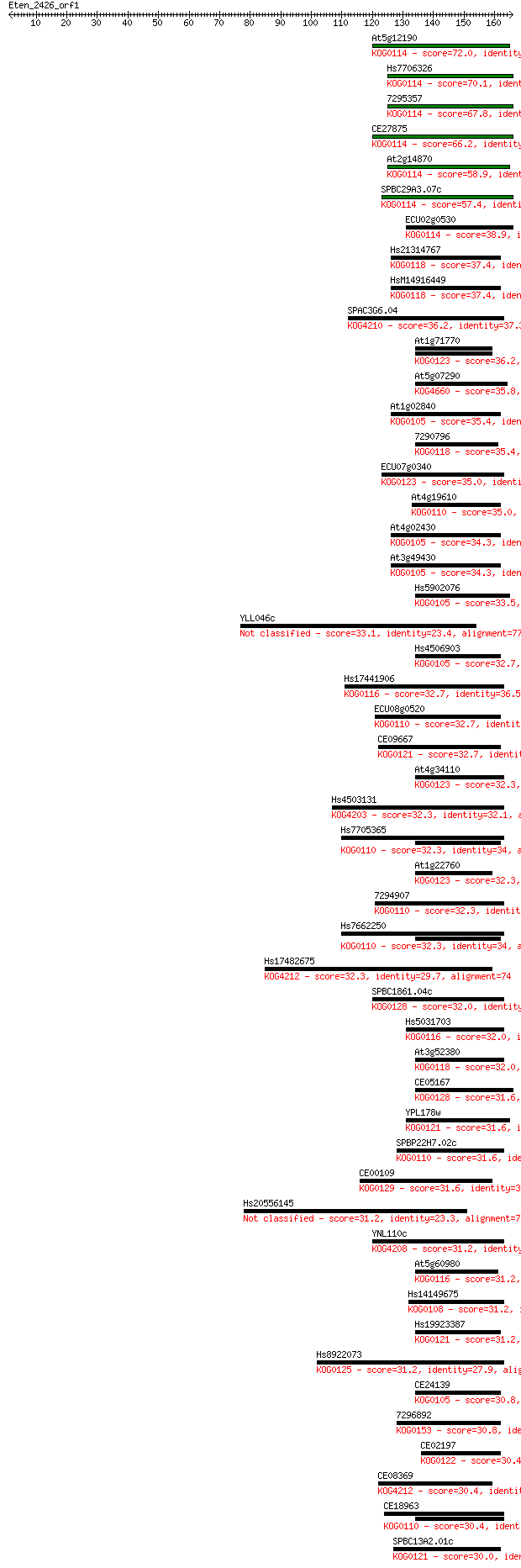
Sequences producing significant alignments: (Bits) Value
At5g12190 72.0 5e-13
Hs7706326 70.1 2e-12
7295357 67.8 9e-12
CE27875 66.2 3e-11
At2g14870 58.9 5e-09
SPBC29A3.07c 57.4 1e-08
ECU02g0530 38.9 0.005
Hs21314767 37.4 0.012
HsM14916449 37.4 0.013
SPAC3G6.04 36.2 0.028
At1g71770 36.2 0.034
At5g07290 35.8 0.039
At1g02840 35.4 0.055
7290796 35.4 0.055
ECU07g0340 35.0 0.059
At4g19610 35.0 0.073
At4g02430 34.3 0.10
At3g49430 34.3 0.11
Hs5902076 33.5 0.19
YLL046c 33.1 0.25
Hs4506903 32.7 0.30
Hs17441906 32.7 0.32
ECU08g0520 32.7 0.33
CE09667 32.7 0.36
At4g34110 32.3 0.39
Hs4503131 32.3 0.42
Hs7705365 32.3 0.42
At1g22760 32.3 0.43
7294907 32.3 0.44
Hs7662250 32.3 0.44
Hs17482675 32.3 0.49
SPBC1861.04c 32.0 0.58
Hs5031703 32.0 0.58
At3g52380 32.0 0.63
CE05167 31.6 0.65
YPL178w 31.6 0.72
SPBP22H7.02c 31.6 0.75
CE00109 31.6 0.79
Hs20556145 31.2 0.91
YNL110c 31.2 0.94
At5g60980 31.2 0.98
Hs14149675 31.2 1.0
Hs19923387 31.2 1.1
Hs8922073 31.2 1.1
CE24139 30.8 1.1
7296892 30.8 1.1
CE02197 30.4 1.5
CE08369 30.4 1.6
CE18963 30.4 1.8
SPBC13A2.01c 30.0 2.0
> At5g12190
Length=124
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 31/45 (68%), Positives = 36/45 (80%), Gaps = 0/45 (0%)
Query 120 RGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRG 164
R RL PEV+R+LYVRNLPF I EE+YD+FGKYG+IRQIR G
Sbjct 7 RKSNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIG 51
> Hs7706326
Length=125
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 30/41 (73%), Positives = 35/41 (85%), Gaps = 0/41 (0%)
Query 125 RLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGT 165
RL PEV+RILY+RNLP+KI EE+YD+FGKYG IRQIR G
Sbjct 12 RLPPEVNRILYIRNLPYKITAEEMYDIFGKYGPIRQIRVGN 52
> 7295357
Length=121
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 28/41 (68%), Positives = 36/41 (87%), Gaps = 0/41 (0%)
Query 125 RLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGT 165
RL PEV+R+LYVRNLP+KI +E+YD+FGK+G+IRQIR G
Sbjct 8 RLPPEVNRLLYVRNLPYKITSDEMYDIFGKFGAIRQIRVGN 48
> CE27875
Length=215
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 26/46 (56%), Positives = 38/46 (82%), Gaps = 0/46 (0%)
Query 120 RGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGT 165
+ R +L PEV+RILY++NLP+KI EE+Y++FGK+G++RQIR G
Sbjct 7 QNRGAKLPPEVNRILYIKNLPYKITTEEMYEIFGKFGAVRQIRVGN 52
> At2g14870
Length=101
Score = 58.9 bits (141), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 24/40 (60%), Positives = 32/40 (80%), Gaps = 0/40 (0%)
Query 125 RLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRG 164
RL PEV+R+LY+ NLPF I E+ YD+FG+Y +IRQ+R G
Sbjct 13 RLPPEVTRLLYICNLPFSITSEDTYDLFGRYSTIRQVRIG 52
> SPBC29A3.07c
Length=137
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 23/43 (53%), Positives = 32/43 (74%), Gaps = 0/43 (0%)
Query 123 PQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGT 165
P + EV+ IL+++NL FKI EE+YD+FG+YG +RQIR G
Sbjct 3 PSTVNQEVNSILFIKNLSFKITAEEMYDLFGRYGPVRQIRLGN 45
> ECU02g0530
Length=93
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 28/35 (80%), Gaps = 0/35 (0%)
Query 131 SRILYVRNLPFKINGEELYDVFGKYGSIRQIRRGT 165
++IL+VRNLP ++ +++ ++FG+YG+I QIR G
Sbjct 6 TQILFVRNLPKDVSKDKVVELFGEYGTIVQIRIGV 40
> Hs21314767
Length=217
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 1/36 (2%)
Query 126 LAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQI 161
LAP S + YV NLPF + +LY +F KYG + ++
Sbjct 5 LAPSKSTV-YVSNLPFSLTNNDLYRIFSKYGKVVKV 39
> HsM14916449
Length=217
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 1/36 (2%)
Query 126 LAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQI 161
LAP S + YV NLPF + +LY +F KYG + ++
Sbjct 5 LAPSKSTV-YVSNLPFSLTNNDLYRIFSKYGKVVKV 39
> SPAC3G6.04
Length=369
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 112 QPAPLLAGRGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIR 162
+PA L+ Q E S IL+V NL F+ +L + FG+ G IR++R
Sbjct 208 KPANTLSKTASIQSSKKEPSSILFVGNLDFETTDADLKEHFGQVGQIRRVR 258
> At1g71770
Length=668
Score = 36.2 bits (82), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 14/25 (56%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSI 158
+YV+NLP +I +EL FGKYG I
Sbjct 227 VYVKNLPKEITDDELKKTFGKYGDI 251
Score = 30.4 bits (67), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 10/25 (40%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSI 158
LY++NL +N E+L ++F +YG++
Sbjct 330 LYLKNLDDSVNDEKLKEMFSEYGNV 354
> At5g07290
Length=907
Score = 35.8 bits (81), Expect = 0.039, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQIRR 163
L+V NL I+ EEL+ +F YG IR++RR
Sbjct 297 LWVNNLDSSISNEELHGIFSSYGEIREVRR 326
> At1g02840
Length=283
Score = 35.4 bits (80), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 126 LAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQI 161
++ SR +YV NLP I E+ D+F KYG + QI
Sbjct 1 MSSRSSRTVYVGNLPGDIREREVEDLFSKYGPVVQI 36
> 7290796
Length=334
Score = 35.4 bits (80), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 16/27 (59%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQ 160
LYV NLP I ++L +FGKYGSI Q
Sbjct 173 LYVTNLPRTITDDQLDTIFGKYGSIVQ 199
> ECU07g0340
Length=333
Score = 35.0 bits (79), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 123 PQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIR 162
P+ L P I+Y+ N+ I E+L D+F K G I+ +R
Sbjct 133 PKELRPPDESIVYISNIDLNIEEEQLEDIFKKMGEIKGVR 172
> At4g19610
Length=772
Score = 35.0 bits (79), Expect = 0.073, Method: Composition-based stats.
Identities = 15/29 (51%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 133 ILYVRNLPFKINGEELYDVFGKYGSIRQI 161
IL V+NLPF +EL +FGK+GS+ +I
Sbjct 410 ILLVKNLPFASTEKELAQMFGKFGSLDKI 438
> At4g02430
Length=294
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 126 LAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQI 161
++ SR +YV NLP I E+ D+F KYG + QI
Sbjct 1 MSSRSSRTIYVGNLPGDIREREVEDLFSKYGPVVQI 36
> At3g49430
Length=243
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 126 LAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQI 161
++ SR +YV NLP I E+ D+F KYG I I
Sbjct 1 MSGRFSRSIYVGNLPGDIREHEIEDIFYKYGRIVDI 36
> Hs5902076
Length=248
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 18/35 (51%), Positives = 23/35 (65%), Gaps = 4/35 (11%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQI----RRG 164
+YV NLP I +++ DVF KYG+IR I RRG
Sbjct 18 IYVGNLPPDIRTKDIEDVFYKYGAIRDIDLKNRRG 52
> YLL046c
Length=249
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 18/81 (22%), Positives = 31/81 (38%), Gaps = 4/81 (4%)
Query 77 FSAFFAGCSLSGVPRECCGAAAAAAAAAAAKSPAMQPAPLLAGRGRPQRLAPEV----SR 132
F F G +L V + G + + ++ AP++ +
Sbjct 81 FIEFQEGVNLKKVKEKMNGKIFMNEKIVIENILTKEEKSFEKNQKSNKKTAPDLKPLSTN 140
Query 133 ILYVRNLPFKINGEELYDVFG 153
LYV+N+P K E+L +FG
Sbjct 141 TLYVKNIPMKSTNEDLAKIFG 161
> Hs4506903
Length=221
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQI 161
+YV NLP + ++L D+F KYG IR+I
Sbjct 16 IYVGNLPTDVREKDLEDLFYKYGRIREI 43
> Hs17441906
Length=334
Score = 32.7 bits (73), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 31/58 (53%), Gaps = 7/58 (12%)
Query 111 MQPAPLLA----GRGRPQRLA--PEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIR 162
M P P+ G PQR+ P+ S L++ NLP +++ EL D F YG++ ++
Sbjct 192 MGPRPICEAGEQGDIEPQRVVRHPD-SHQLFIGNLPHEVDKSELKDFFQNYGNVVELH 248
> ECU08g0520
Length=432
Score = 32.7 bits (73), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 13/41 (31%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 121 GRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQI 161
G + + SR +++RN+P + N + + DVF +YG I ++
Sbjct 103 GESEERMIKYSRKIFIRNVPAEANEQFVRDVFKEYGEIEEV 143
> CE09667
Length=154
Score = 32.7 bits (73), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 122 RPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQI 161
R Q A S LYV NL + +++Y++FG+ G +R++
Sbjct 27 RDQETALRTSCTLYVGNLSYYTKEDQVYELFGRAGDVRRV 66
> At4g34110
Length=629
Score = 32.3 bits (72), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQIR 162
LYV +L F + +L+D FG+ G++ +R
Sbjct 38 LYVGDLDFNVTDSQLFDAFGQMGTVVTVR 66
> Hs4503131
Length=781
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 28/58 (48%), Gaps = 2/58 (3%)
Query 107 KSPAMQPAPLLAGRGRPQRLAPEVSRILYVRNLPF--KINGEELYDVFGKYGSIRQIR 162
S A AP L+G+G P+ + S++LY F E++ D+ G+Y R R
Sbjct 36 HSGATTTAPSLSGKGNPEEEDVDTSQVLYEWEQGFSQSFTQEQVADIDGQYAMTRAQR 93
> Hs7705365
Length=960
Score = 32.3 bits (72), Expect = 0.42, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 32/53 (60%), Gaps = 2/53 (3%)
Query 110 AMQPAPLLAGRGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIR 162
A +PA LA + + R S+IL VRN+PF+ + E+ ++F +G ++ +R
Sbjct 812 ATKPAVTLARKKQVPR-KQTTSKIL-VRNIPFQAHSREIRELFSTFGELKTVR 862
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQI 161
L+VRNLP+ E+L +F KYG + ++
Sbjct 404 LFVRNLPYTSTEEDLEKLFSKYGPLSEL 431
> At1g22760
Length=660
Score = 32.3 bits (72), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 13/25 (52%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSI 158
+YV+NLP +I +EL FGK+G I
Sbjct 231 VYVKNLPKEIGEDELRKTFGKFGVI 255
> 7294907
Length=918
Score = 32.3 bits (72), Expect = 0.44, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 26/45 (57%), Gaps = 3/45 (6%)
Query 121 GRPQRLAPEVSRI---LYVRNLPFKINGEELYDVFGKYGSIRQIR 162
G +RLA + + + VRN+PF+ E+ D+F +G +R +R
Sbjct 771 GAQRRLASQKKQTGTKILVRNIPFQAQYREVRDIFKAFGELRSLR 815
> Hs7662250
Length=960
Score = 32.3 bits (72), Expect = 0.44, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 32/53 (60%), Gaps = 2/53 (3%)
Query 110 AMQPAPLLAGRGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIR 162
A +PA LA + + R S+IL VRN+PF+ + E+ ++F +G ++ +R
Sbjct 812 ATKPAVTLARKKQVPR-KQTTSKIL-VRNIPFQAHSREIRELFSTFGELKTVR 862
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQI 161
L+VRNLP+ E+L +F KYG + ++
Sbjct 404 LFVRNLPYTSTEEDLEKLFSKYGPLSEL 431
> Hs17482675
Length=175
Score = 32.3 bits (72), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 35/75 (46%), Gaps = 8/75 (10%)
Query 85 SLSGVPRECCGAAAAA-AAAAAAKSPAMQPAPLLAGRGRPQRLAPEVSRILYVRNLPFKI 143
SL+ + CG+ A A A + P ++ +L G R + ++VRNLPF
Sbjct 66 SLTCMGLAMCGSGGATFDHAIALQVPLVEQETMLLGMARK-------ACQIFVRNLPFNF 118
Query 144 NGEELYDVFGKYGSI 158
+ L D F +YG +
Sbjct 119 TWKMLKDKFNEYGHM 133
> SPBC1861.04c
Length=1014
Score = 32.0 bits (71), Expect = 0.58, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 120 RGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIR 162
R P+ A R LYV N+ FK+N +++ F YG + +R
Sbjct 745 RRTPRSGAVYEGRELYVTNIDFKVNEKDVETFFRDYGQVESVR 787
> Hs5031703
Length=466
Score = 32.0 bits (71), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 131 SRILYVRNLPFKINGEELYDVFGKYGSIRQIR 162
S L++ NLP +++ EL D F YG++ ++R
Sbjct 339 SHQLFIGNLPHEVDKSELKDFFQSYGNVVELR 370
> At3g52380
Length=329
Score = 32.0 bits (71), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQIR 162
LYV NLP+ I EL +FG+ G++ ++
Sbjct 118 LYVGNLPYTITSSELSQIFGEAGTVVDVQ 146
> CE05167
Length=836
Score = 31.6 bits (70), Expect = 0.65, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQIRRGT 165
++VRN+ F+ +EL +F K+G++ +RR T
Sbjct 685 VFVRNVHFQATDDELKALFSKFGTVTSVRRVT 716
> YPL178w
Length=208
Score = 31.6 bits (70), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 131 SRILYVRNLPFKINGEELYDVFGKYGSIRQIRRG 164
S +YV NL F + E++Y++F K G+I++I G
Sbjct 45 SSTIYVGNLSFYTSEEQIYELFSKCGTIKRIIMG 78
> SPBP22H7.02c
Length=833
Score = 31.6 bits (70), Expect = 0.75, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 24/35 (68%), Gaps = 1/35 (2%)
Query 128 PEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIR 162
P+ ++IL ++NLPF+ +++ + G YG +R +R
Sbjct 718 PKGTKIL-IKNLPFEATKKDVQSLLGAYGQLRSVR 751
> CE00109
Length=372
Score = 31.6 bits (70), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 116 LLAGRGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSI 158
L A + RP P SR +++ LP ++ E+++ FG +G +
Sbjct 42 LEASKKRPISAEPLYSRKVFIGGLPIDVSEEQVWATFGAFGKV 84
> Hs20556145
Length=152
Score = 31.2 bits (69), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 17/73 (23%), Positives = 27/73 (36%), Gaps = 0/73 (0%)
Query 78 SAFFAGCSLSGVPRECCGAAAAAAAAAAAKSPAMQPAPLLAGRGRPQRLAPEVSRILYVR 137
S + GC++ G ++C A A + PL + P + +
Sbjct 57 SGYSYGCAVDGNGKDCFSAHETPEHTAGTLVMPKETTPLAENQDEDPLEDPHLHLNIEES 116
Query 138 NLPFKINGEELYD 150
N F + EELYD
Sbjct 117 NQEFMVKSEELYD 129
> YNL110c
Length=220
Score = 31.2 bits (69), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 120 RGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIR 162
+ + ++ E S I+YV LP + +EL F ++G ++++R
Sbjct 79 KSKDKKTLEEYSGIIYVSRLPHGFHEKELSKYFAQFGDLKEVR 121
> At5g60980
Length=459
Score = 31.2 bits (69), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQ 160
+YVRNLPF +L +VF +G+I+
Sbjct 295 IYVRNLPFDSTPTQLEEVFKNFGAIKH 321
> Hs14149675
Length=616
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 132 RILYVRNLPFKINGEELYDVFGKYGSIRQIR 162
R ++V N+P++ E+L D+F + GS+ R
Sbjct 16 RSVFVGNIPYEATEEQLKDIFSEVGSVVSFR 46
> Hs19923387
Length=156
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQI 161
LYV NL F E++Y++F K G I++I
Sbjct 42 LYVGNLSFYTTEEQIYELFSKSGDIKKI 69
> Hs8922073
Length=377
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 29/61 (47%), Gaps = 8/61 (13%)
Query 102 AAAAAKSPAMQPAPLLAGRGRPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQI 161
AA P QP+ + +P+RL +V N+PF+ +L +FG++G I +
Sbjct 75 AAPTDGQPQTQPSENTENKSQPKRL--------HVSNIPFRFRDPDLRQMFGQFGKILDV 126
Query 162 R 162
Sbjct 127 E 127
> CE24139
Length=235
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQI 161
+YV NLP + +E+ D+F KYG I+ +
Sbjct 11 VYVGNLPGDVREKEVEDIFHKYGRIKYV 38
> 7296892
Length=416
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 18/36 (50%), Positives = 21/36 (58%), Gaps = 2/36 (5%)
Query 128 PEVSRI--LYVRNLPFKINGEELYDVFGKYGSIRQI 161
PE I LYV NLP +I EL D F ++G IR I
Sbjct 224 PEDRNITTLYVGNLPEEITEPELRDQFYQFGEIRSI 259
> CE02197
Length=256
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 136 VRNLPFKINGEELYDVFGKYGSIRQI 161
V NLP ++N +EL D+FGK G + +I
Sbjct 180 VTNLPQEMNEDELRDLFGKIGRVIRI 205
> CE08369
Length=454
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 122 RPQRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSI 158
R + P + +++ NL F + ++LY+VFG G I
Sbjct 178 RQHNIEPPLCERIFIANLAFNVGTDKLYEVFGMAGKI 214
> CE18963
Length=872
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 27/39 (69%), Gaps = 1/39 (2%)
Query 124 QRLAPEVSRILYVRNLPFKINGEELYDVFGKYGSIRQIR 162
Q+ E +++L VRNLPF+ + +E+ +F +G+++ IR
Sbjct 741 QKEQGECTKLL-VRNLPFEASVKEVETLFETFGAVKTIR 778
Score = 29.3 bits (64), Expect = 3.9, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 134 LYVRNLPFKINGEELYDVFGKYGSIRQIR 162
L++RNLP+ ++L +F KYG + +++
Sbjct 281 LFLRNLPYATKEDDLQFLFKKYGEVSEVQ 309
> SPBC13A2.01c
Length=182
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 127 APEVSRILYVRNLPFKINGEELYDVFGKYGSIRQI 161
A + S +YV NL F E++Y +F K G IR+I
Sbjct 27 AVKQSNCVYVGNLSFYTTEEQIYALFSKCGEIRRI 61
Lambda K H
0.323 0.137 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2353551590
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40