bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2421_orf2
Length=135
Score E
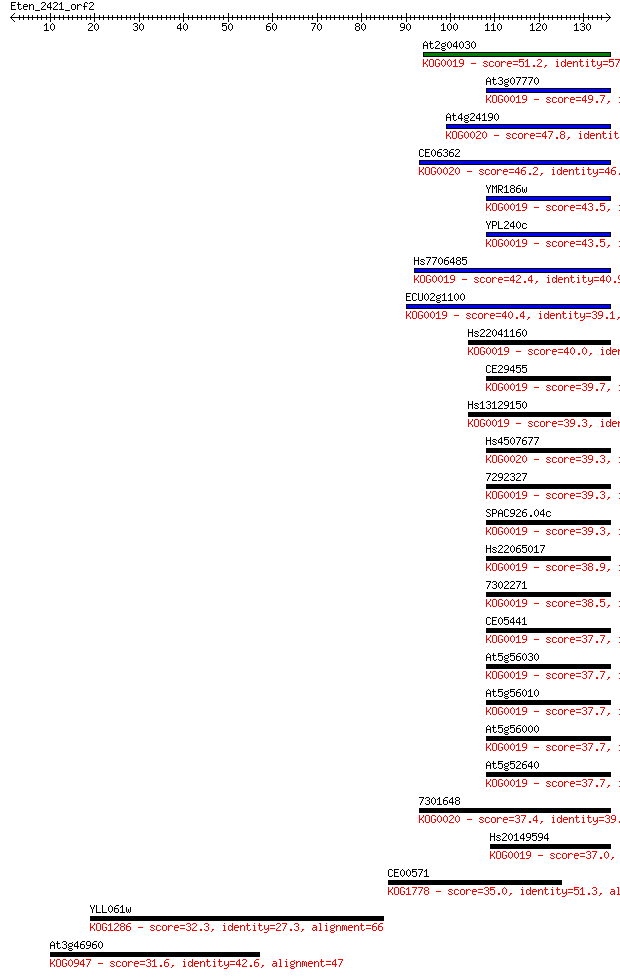
Sequences producing significant alignments: (Bits) Value
At2g04030 51.2 5e-07
At3g07770 49.7 2e-06
At4g24190 47.8 6e-06
CE06362 46.2 2e-05
YMR186w 43.5 1e-04
YPL240c 43.5 1e-04
Hs7706485 42.4 2e-04
ECU02g1100 40.4 0.001
Hs22041160 40.0 0.001
CE29455 39.7 0.002
Hs13129150 39.3 0.002
Hs4507677 39.3 0.002
7292327 39.3 0.002
SPAC926.04c 39.3 0.002
Hs22065017 38.9 0.002
7302271 38.5 0.003
CE05441 37.7 0.005
At5g56030 37.7 0.006
At5g56010 37.7 0.006
At5g56000 37.7 0.006
At5g52640 37.7 0.006
7301648 37.4 0.009
Hs20149594 37.0 0.009
CE00571 35.0 0.042
YLL061w 32.3 0.27
At3g46960 31.6 0.41
> At2g04030
Length=780
Score = 51.2 bits (121), Expect = 5e-07, Method: Composition-based stats.
Identities = 24/42 (57%), Positives = 31/42 (73%), Gaps = 0/42 (0%)
Query 94 AELAKLRADAEGHQETHEYQAEVTRLMDIIVNSLYSQREVFL 135
A +A+ EG E EYQAEV+RL+D+IV+SLYS +EVFL
Sbjct 63 AAVAEKETTEEGSGEKFEYQAEVSRLLDLIVHSLYSHKEVFL 104
> At3g07770
Length=803
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/28 (78%), Positives = 25/28 (89%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
E EYQAEV+RLMD+IVNSLYS +EVFL
Sbjct 95 EKFEYQAEVSRLMDLIVNSLYSNKEVFL 122
> At4g24190
Length=823
Score = 47.8 bits (112), Expect = 6e-06, Method: Composition-based stats.
Identities = 22/37 (59%), Positives = 30/37 (81%), Gaps = 4/37 (10%)
Query 99 LRADAEGHQETHEYQAEVTRLMDIIVNSLYSQREVFL 135
LR++AE E+QAEV+RLMDII+NSLYS +++FL
Sbjct 72 LRSNAE----KFEFQAEVSRLMDIIINSLYSNKDIFL 104
> CE06362
Length=760
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 20/43 (46%), Positives = 32/43 (74%), Gaps = 4/43 (9%)
Query 93 LAELAKLRADAEGHQETHEYQAEVTRLMDIIVNSLYSQREVFL 135
++++ +LR+ AE HE+QAEV R+M +I+NSLY +E+FL
Sbjct 51 VSQIKELRSKAE----KHEFQAEVNRMMKLIINSLYRNKEIFL 89
> YMR186w
Length=705
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 16/28 (57%), Positives = 25/28 (89%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
ET E+QAE+T+LM +I+N++YS +E+FL
Sbjct 4 ETFEFQAEITQLMSLIINTVYSNKEIFL 31
> YPL240c
Length=709
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 16/28 (57%), Positives = 25/28 (89%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
ET E+QAE+T+LM +I+N++YS +E+FL
Sbjct 4 ETFEFQAEITQLMSLIINTVYSNKEIFL 31
> Hs7706485
Length=704
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 27/44 (61%), Gaps = 0/44 (0%)
Query 92 PLAELAKLRADAEGHQETHEYQAEVTRLMDIIVNSLYSQREVFL 135
PL + +G HE+QAE +L+DI+ SLYS++EVF+
Sbjct 70 PLHSIISSTESVQGSTSKHEFQAETKKLLDIVARSLYSEKEVFI 113
> ECU02g1100
Length=690
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 18/46 (39%), Positives = 31/46 (67%), Gaps = 4/46 (8%)
Query 90 QGPLAELAKLRADAEGHQETHEYQAEVTRLMDIIVNSLYSQREVFL 135
Q PL E K++ + H ETH ++ +V ++MD ++ S+YS +E+FL
Sbjct 5 QEPLVE-GKIK---DKHSETHGFEVDVNQMMDTMIKSVYSSKELFL 46
> Hs22041160
Length=343
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 104 EGHQETHEYQAEVTRLMDIIVNSLYSQREVFL 135
E ET +QAE+ +LM +I+N+ YS +E+FL
Sbjct 9 EKEVETFAFQAEIAQLMSLIINTFYSNKEIFL 40
> CE29455
Length=657
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 16/28 (57%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
+ HE+QAE LMDI+ SLYS EVF+
Sbjct 28 QRHEFQAETRNLMDIVAKSLYSHSEVFV 55
> Hs13129150
Length=732
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 104 EGHQETHEYQAEVTRLMDIIVNSLYSQREVFL 135
E ET +QAE+ +LM +I+N+ YS +E+FL
Sbjct 14 EEEVETFAFQAEIAQLMSLIINTFYSNKEIFL 45
> Hs4507677
Length=803
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
E +QAEV R+M +I+NSLY +E+FL
Sbjct 74 EKFAFQAEVNRMMKLIINSLYKNKEIFL 101
> 7292327
Length=717
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
ET +QAE+ +LM +I+N+ YS +E+FL
Sbjct 6 ETFAFQAEIAQLMSLIINTFYSNKEIFL 33
> SPAC926.04c
Length=704
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 25/28 (89%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
ET +++AE+++LM +I+N++YS +E+FL
Sbjct 5 ETFKFEAEISQLMSLIINTVYSNKEIFL 32
> Hs22065017
Length=343
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
ET +QAE+ +LM +I+N+ YS +E+FL
Sbjct 18 ETFAFQAEIAQLMSLIINTFYSNKEIFL 45
> 7302271
Length=691
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 15/28 (53%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
+ HE+QAE +L+DI+ SLYS EVF+
Sbjct 64 DKHEFQAETRQLLDIVARSLYSDHEVFV 91
> CE05441
Length=702
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
ET +QAE+ +LM +I+N+ YS +E++L
Sbjct 6 ETFAFQAEIAQLMSLIINTFYSNKEIYL 33
> At5g56030
Length=699
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
ET +QAE+ +L+ +I+N+ YS +E+FL
Sbjct 5 ETFAFQAEINQLLSLIINTFYSNKEIFL 32
> At5g56010
Length=699
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
ET +QAE+ +L+ +I+N+ YS +E+FL
Sbjct 5 ETFAFQAEINQLLSLIINTFYSNKEIFL 32
> At5g56000
Length=699
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
ET +QAE+ +L+ +I+N+ YS +E+FL
Sbjct 5 ETFAFQAEINQLLSLIINTFYSNKEIFL 32
> At5g52640
Length=705
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 108 ETHEYQAEVTRLMDIIVNSLYSQREVFL 135
ET +QAE+ +L+ +I+N+ YS +E+FL
Sbjct 10 ETFAFQAEINQLLSLIINTFYSNKEIFL 37
> 7301648
Length=787
Score = 37.4 bits (85), Expect = 0.009, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 28/43 (65%), Gaps = 4/43 (9%)
Query 93 LAELAKLRADAEGHQETHEYQAEVTRLMDIIVNSLYSQREVFL 135
+A+L ++R A+ +Q EV R+M +I+NSLY +E+FL
Sbjct 62 VAQLKEIREKAK----KFTFQTEVNRMMKLIINSLYRNKEIFL 100
> Hs20149594
Length=724
Score = 37.0 bits (84), Expect = 0.009, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 21/27 (77%), Gaps = 0/27 (0%)
Query 109 THEYQAEVTRLMDIIVNSLYSQREVFL 135
T +QAE+ +LM +I+N+ YS +E+FL
Sbjct 14 TFAFQAEIAQLMSLIINTFYSNKEIFL 40
> CE00571
Length=322
Score = 35.0 bits (79), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 23/46 (50%), Gaps = 7/46 (15%)
Query 86 QGAPQGPLAELAKLRADAEGHQETHEYQAEVTR-------LMDIIV 124
QGA Q PL E A++R D GHQ + E R LMDI V
Sbjct 250 QGAAQSPLDEFARMRIDEGGHQLRTNQETETNRQNQSQQPLMDINV 295
> YLL061w
Length=583
Score = 32.3 bits (72), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 18/67 (26%), Positives = 35/67 (52%), Gaps = 1/67 (1%)
Query 19 SSSSRRSAATVSSILKMALWRVVPFCFLGLLLLRCSA-AADPAGASDAAAAEPAAAPAAA 77
+S+ RS ++VS K WR++ F + ++++ C DP S +++ + A+P
Sbjct 283 TSTEARSVSSVSRAAKGTFWRIIIFYIVTVIIIGCLVPYNDPRLISGSSSEDITASPFVI 342
Query 78 AAAAEGA 84
A + GA
Sbjct 343 ALSNTGA 349
> At3g46960
Length=1347
Score = 31.6 bits (70), Expect = 0.41, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 30/52 (57%), Gaps = 5/52 (9%)
Query 10 SSCGGSSSSSSSSRRSAAT-----VSSILKMALWRVVPFCFLGLLLLRCSAA 56
SS G+S ++ + RRSAA+ ++ + KM+L VV FCF RC+ A
Sbjct 608 SSYSGNSQNNGAFRRSAASNWLLLINKLSKMSLLPVVVFCFSKNYCDRCADA 659
Lambda K H
0.313 0.122 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1388795348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40