bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
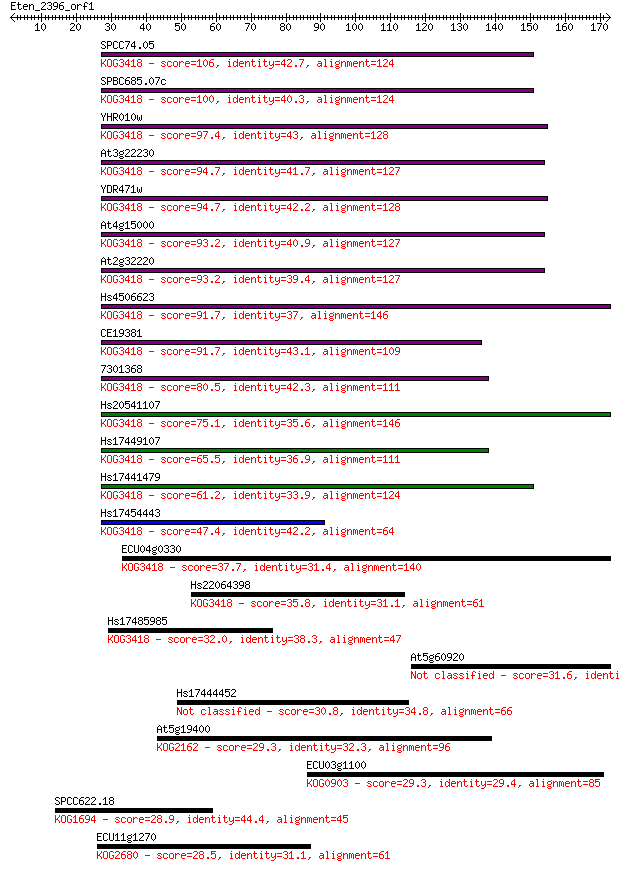
Query= Eten_2396_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
SPCC74.05 106 3e-23
SPBC685.07c 100 2e-21
YHR010w 97.4 1e-20
At3g22230 94.7 7e-20
YDR471w 94.7 7e-20
At4g15000 93.2 2e-19
At2g32220 93.2 2e-19
Hs4506623 91.7 6e-19
CE19381 91.7 6e-19
7301368 80.5 2e-15
Hs20541107 75.1 6e-14
Hs17449107 65.5 5e-11
Hs17441479 61.2 1e-09
Hs17454443 47.4 1e-05
ECU04g0330 37.7 0.010
Hs22064398 35.8 0.039
Hs17485985 32.0 0.70
At5g60920 31.6 0.84
Hs17444452 30.8 1.5
At5g19400 29.3 3.6
ECU03g1100 29.3 4.1
SPCC622.18 28.9 5.1
ECU11g1270 28.5 7.3
> SPCC74.05
Length=136
Score = 106 bits (264), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 53/125 (42%), Positives = 80/125 (64%), Gaps = 2/125 (1%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
MVK+LKPG+V +V GR AGKK V++ + +G+K F + +VAG+E+ PL+V+K M
Sbjct 1 MVKILKPGKVALVTRGRFAGKKVVILQNVDQGSKSHPFGHAVVAGVERYPLKVTKSMGAK 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFY 145
++ KR R K FI+ +N HLMPTRY + D ++K L + + + R +AKK + N F
Sbjct 61 RIAKRSRVKPFIKVINYNHLMPTRYALELD-NLKGLVTPTTFSEPSQRSAAKKTVKNTFE 119
Query 146 EKFMN 150
EK+
Sbjct 120 EKYQT 124
> SPBC685.07c
Length=136
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 50/125 (40%), Positives = 80/125 (64%), Gaps = 2/125 (1%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
MVK+LKPG+V ++ GR AGKK V++ + +G+K F + +VAG+E+ PL+V+K M
Sbjct 1 MVKILKPGKVALITRGRFAGKKVVILQAIDQGSKSHPFGHAVVAGVERYPLKVTKSMGAK 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFY 145
++ +R R K FI+ VN HLMPTRY + D ++K L + ++ R +A+K + F
Sbjct 61 RIARRSRVKPFIKVVNYNHLMPTRYALELD-NLKGLITADTFKEPTQRSAARKTVKKTFE 119
Query 146 EKFMN 150
EK+ +
Sbjct 120 EKYQS 124
> YHR010w
Length=136
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 55/129 (42%), Positives = 77/129 (59%), Gaps = 2/129 (1%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVVS-TSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
M K LK G+V VV+ GR AGKK V+V EG+K F + LVAGIE+ PL+V+K+
Sbjct 1 MAKFLKAGKVAVVVRGRYAGKKVVIVKPHDEGSKSHPFGHALVAGIERYPLKVTKKHGAK 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFY 145
KV KR + K FI+ VN HL+PTRYT+ + K++ + E + R+ AKK + F
Sbjct 61 KVAKRTKIKPFIKVVNYNHLLPTRYTLDVE-AFKSVVSTETFEQPSQREEAKKVVKKAFE 119
Query 146 EKFMNPVNE 154
E+ N+
Sbjct 120 ERHQAGKNQ 128
> At3g22230
Length=135
Score = 94.7 bits (234), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 53/128 (41%), Positives = 78/128 (60%), Gaps = 3/128 (2%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
MVK LK + V++L GR AGKK V++ S +GT +R + +CLVAG++K P +V ++ S
Sbjct 1 MVKFLKQNKAVILLQGRYAGKKAVIIKSFDDGTSDRRYGHCLVAGLKKYPSKVIRKDSAK 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFY 145
K K+ R K FI+ VN +HLMPTRYT+ D D+K +A +L+ + +A K
Sbjct 61 KTAKKSRVKCFIKLVNYQHLMPTRYTL--DVDLKEVATLDALKSKDKKVTALKEAKAKLE 118
Query 146 EKFMNPVN 153
E+F N
Sbjct 119 ERFKTGKN 126
> YDR471w
Length=136
Score = 94.7 bits (234), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 54/129 (41%), Positives = 76/129 (58%), Gaps = 2/129 (1%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVVS-TSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
M K LK G+V VV+ GR AGKK V+V EG+K F + LVAGIE+ P +V+K+
Sbjct 1 MAKFLKAGKVAVVVRGRYAGKKVVIVKPHDEGSKSHPFGHALVAGIERYPSKVTKKHGAK 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFY 145
KV KR + K FI+ VN HL+PTRYT+ + K++ + E + R+ AKK + F
Sbjct 61 KVAKRTKIKPFIKVVNYNHLLPTRYTLDVE-AFKSVVSTETFEQPSQREEAKKVVKKAFE 119
Query 146 EKFMNPVNE 154
E+ N+
Sbjct 120 ERHQAGKNQ 128
> At4g15000
Length=135
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 52/128 (40%), Positives = 78/128 (60%), Gaps = 3/128 (2%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
MVK LK + V++L GR AGKK V++ S +G ++R + +CLVAG++K P +V ++ S
Sbjct 1 MVKFLKQNKAVILLQGRYAGKKAVIIKSFDDGNRDRPYGHCLVAGLKKYPSKVIRKDSAK 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFY 145
K K+ R K FI+ VN +HLMPTRYT+ D D+K +A +L+ + +A K
Sbjct 61 KTAKKSRVKCFIKLVNYQHLMPTRYTL--DVDLKEVATLDALQSKDKKVAALKEAKAKLE 118
Query 146 EKFMNPVN 153
E+F N
Sbjct 119 ERFKTGKN 126
> At2g32220
Length=135
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 50/128 (39%), Positives = 77/128 (60%), Gaps = 3/128 (2%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
MVK +KPG+ V++L GR GKK V+V S +GT E+ + +CLVAG++K P +V ++ S
Sbjct 1 MVKCMKPGKAVILLQGRYTGKKAVIVKSFDDGTVEKKYGHCLVAGLKKYPSKVIRKDSAK 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFY 145
K K+ R K F + +N +H+MPTRYT+ D D+K + ++ + +A K F
Sbjct 61 KTAKKSRVKCFFKVINYQHVMPTRYTL--DLDLKNVVSADAISSKDKKVTALKEAKAKFE 118
Query 146 EKFMNPVN 153
E+F N
Sbjct 119 ERFKTGKN 126
> Hs4506623
Length=136
Score = 91.7 bits (226), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 54/147 (36%), Positives = 86/147 (58%), Gaps = 12/147 (8%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
M K +KPG+VV+VL GR +G+K V+V + +GT +R + + LVAGI++ P +V+ M
Sbjct 1 MGKFMKPGKVVLVLAGRYSGRKAVIVKNIDDGTSDRPYSHALVAGIDRYPRKVTAAMGKK 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFY 145
K+ KR + K+F++ N HLMPTRY+V D K + + D A+++ A++ F
Sbjct 61 KIAKRSKIKSFVKVYNYNHLMPTRYSVDIPLD-KTVVNKDVFRDPALKRKARREAKVKFE 119
Query 146 EKFMNPVNEKAGKVSKDVLFLRKKLRF 172
E++ K K+ F +KLRF
Sbjct 120 ERY---------KTGKNKWFF-QKLRF 136
> CE19381
Length=136
Score = 91.7 bits (226), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 47/110 (42%), Positives = 72/110 (65%), Gaps = 1/110 (0%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
M K++KPG+VV+VL G+ AG+K VVV EG +R + + ++AGI++ PL+V+K M
Sbjct 1 MGKIMKPGKVVLVLRGKYAGRKAVVVKQQDEGVSDRTYPHAIIAGIDRYPLKVTKDMGKK 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKS 135
K++KR + K F++ V+ HL+PTRY+V FD + EA S RK+
Sbjct 61 KIEKRNKLKPFLKVVSYTHLLPTRYSVDVAFDKTNINKEALKAPSKKRKA 110
> 7301368
Length=135
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 47/112 (41%), Positives = 79/112 (70%), Gaps = 3/112 (2%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVVST-SEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
M K++K G++V+VL+GR AG+K ++V T +GT E+ F + LVAGI++ P +V+K+M
Sbjct 1 MRKIMKQGKIVIVLSGRYAGRKAIIVKTHDDGTPEKPFGHALVAGIDRYPRKVTKKMGKN 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAK 137
K++K+ + K F++ +N HLMPTRYT ++D + L+P+ L+D RK+ +
Sbjct 61 KLKKKSKVKPFLKSLNYNHLMPTRYT-AHDISFEKLSPK-DLKDPVKRKTHR 110
> Hs20541107
Length=132
Score = 75.1 bits (183), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 52/147 (35%), Positives = 82/147 (55%), Gaps = 16/147 (10%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
M K +KPG+VV+VL G AG+K ++V + +GT + F + LVAGI++ PL+V+ M
Sbjct 1 MGKFMKPGKVVLVLAGGYAGRKAIIVKNNDDGTSDSPFSHALVAGIDRYPLKVTAAMGKK 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFY 145
K+ KR + K+F++ N HLM TRY+V K + + D A+++ A + F
Sbjct 61 KISKRSKIKSFVKVYNYNHLMATRYSVD-----KTVINKDVFRDPALKRKAPREAKVKFE 115
Query 146 EKFMNPVNEKAGKVSKDVLFLRKKLRF 172
E++ K K+ F + KLRF
Sbjct 116 ERY---------KTGKNKWFFQ-KLRF 132
> Hs17449107
Length=111
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 41/112 (36%), Positives = 66/112 (58%), Gaps = 2/112 (1%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
M K +KPG VV+VL G +G K ++V + +GT +R + + LVAGI++ P +V+ M
Sbjct 1 MGKFMKPGTVVLVLAGCYSGCKAIIVKNIDDGTLDRPYSHALVAGIDRCPHKVTAAMGKK 60
Query 86 KVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAK 137
K+ KR + K+F++ N LMPTRY+V D K + + D ++ A+
Sbjct 61 KISKRSKIKSFVKVYNHNQLMPTRYSVDIPLD-KTVINKDVFRDPTLKHKAR 111
> Hs17441479
Length=314
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 42/129 (32%), Positives = 67/129 (51%), Gaps = 7/129 (5%)
Query 27 MVKLLKPGRVVVV----LNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKR 81
M K +KPG+VV+V G +G K ++V + +GT E LVAGI+ P +V+
Sbjct 1 MGKFIKPGKVVLVQARHYTGCYSGCKTIIVKNIDDGTLECPVSCSLVAGIDCYPCKVTAA 60
Query 82 MSTTKVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALA 141
M Q R + K+F++ N HLMPTR++V D K + + D A++ A++
Sbjct 61 MGKKSTQ-RSKTKSFVKVYNYNHLMPTRHSVDTPLD-KTVINKDVFRDPALKHKAQRKAK 118
Query 142 NIFYEKFMN 150
E+ +N
Sbjct 119 VKIKERLIN 127
> Hs17454443
Length=117
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Query 27 MVKLLKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
M K LKP +VV VL G + K V+V + +GT +R + + LVAGI+ P +V M
Sbjct 1 MGKFLKPRKVVFVLAGCYSRHKAVIVKNIDDGTSDRPYSHALVAGIDHYPCKVRAAMGKK 60
Query 86 KVQKR 90
K+ KR
Sbjct 61 KITKR 65
> ECU04g0330
Length=126
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 44/142 (30%), Positives = 59/142 (41%), Gaps = 22/142 (15%)
Query 33 PGRVVVVLNGRMAGKKGVVVSTSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQKRMR 92
P +VVV +G+ AGKK VV+ + ER+ V I K P K KR
Sbjct 5 PNMIVVVTSGKNAGKKAVVIKQID---ERYVLTACVVRIPKEP-----EDHQPKWIKRRN 56
Query 93 PKAFI--RYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFMN 150
K +I + N+ HL+ TRY D + A + +ED+A KK L N + M
Sbjct 57 AKFYIILKKYNIAHLIATRYKA--DLGL-ATIDYSGIEDNA---ETKKVLTNKTKDAMMK 110
Query 151 PVNEKAGKVSKDVLFLRKKLRF 172
E K FL L F
Sbjct 111 AWKENKAK------FLFSSLSF 126
> Hs22064398
Length=102
Score = 35.8 bits (81), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 32/61 (52%), Gaps = 5/61 (8%)
Query 53 STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQKRMRPKAFIRYVNVRHLMPTRYTV 112
+T +GT +R + + LVAG + P +++ + K++K R N HLMP R +V
Sbjct 3 NTDDGTSDRPYSHALVAGFDHYPHKLTAVVGKKKMKKSQRS-----IYNYYHLMPIRCSV 57
Query 113 S 113
Sbjct 58 D 58
> Hs17485985
Length=123
Score = 32.0 bits (71), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 1/48 (2%)
Query 29 KLLKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAP 75
K +KPG+VV+VL +G K V + + +GT + + L+AGI+ P
Sbjct 74 KFMKPGKVVLVLAECYSGHKAVTMKNVDKGTSIFPYSHALMAGIDCYP 121
> At5g60920
Length=391
Score = 31.6 bits (70), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 27/57 (47%), Gaps = 7/57 (12%)
Query 116 FDVKALAPEASLEDSAMRKSAKKALANIFYEKFMNPVNEKAGKVSKDVLFLRKKLRF 172
F+ K+L P A L D+AM K FY F++ G V ++LF + + F
Sbjct 285 FNYKSLTPYAGLNDTAMLWGVK------FYNDFLSEAGP-LGNVQSEILFRKDQSTF 334
> Hs17444452
Length=249
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 39/78 (50%), Gaps = 15/78 (19%)
Query 49 GVVVSTSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQKRMR-----PKAFIR----- 98
G V+T + K +H+C C +A + K L K+ S KVQ+ M+ P F+R
Sbjct 33 GWNVNTGDKPKLQHYCDCTLARLHKEDL---KQRSLNKVQEVMQKPNESPSEFMRCIFKV 89
Query 99 --YVNVRHLMPTRYTVSN 114
Y+++ ++ T + SN
Sbjct 90 FEYLSIINMTFTSQSASN 107
> At5g19400
Length=1092
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 23/100 (23%)
Query 43 RMAGK---KGVVVSTSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQKRMRPKAF-IR 98
R+ GK KG +S + T LVAG EK + ++ M KAF IR
Sbjct 281 RLTGKGRGKGADISLKDAT--------LVAGPEKDKVTIANEML----------KAFSIR 322
Query 99 YVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKK 138
+V++ ++ TR ++ FDV A + +SL + SAK+
Sbjct 323 FVHLNGILFTRTSLETFFDVLA-STSSSLREVISLGSAKE 361
> ECU03g1100
Length=711
Score = 29.3 bits (64), Expect = 4.1, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 44/89 (49%), Gaps = 8/89 (8%)
Query 86 KVQKRMRPKAF---IRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKA-LA 141
K+ + +R + I+ +N H +P + VS F A +E+S + SA+ +
Sbjct 233 KLPRHLRQRGLEMEIKLLN--HNLPAK--VSLPFHSGRFVLAARIEESFLLDSAENSPFL 288
Query 142 NIFYEKFMNPVNEKAGKVSKDVLFLRKKL 170
+F F P +AG+ KD LFL+++L
Sbjct 289 VVFETAFEIPRKSRAGEEIKDALFLKQQL 317
> SPCC622.18
Length=195
Score = 28.9 bits (63), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 27/48 (56%), Gaps = 3/48 (6%)
Query 14 RLDLAKFANKVDKMVKL---LKPGRVVVVLNGRMAGKKGVVVSTSEGT 58
R ++ K A K + KL L PG V ++L GR GK+ VV+S E T
Sbjct 32 RENVPKKARKAVRPTKLRASLAPGTVCILLAGRFRGKRVVVLSQLEDT 79
> ECU11g1270
Length=418
Score = 28.5 bits (62), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Query 26 KMVKLLKPGRVVVVLNGRMAGKKGVVVSTSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT 85
KMV+ K G+VV++ R +GK + + S+ HF ++G E L +SK + T
Sbjct 50 KMVESNKGGKVVLIKGDRGSGKTALAIGLSKSLGGVHFNS--ISGTEIYSLEMSKSEAIT 107
Query 86 K 86
+
Sbjct 108 Q 108
Lambda K H
0.323 0.136 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2562785186
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40