bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
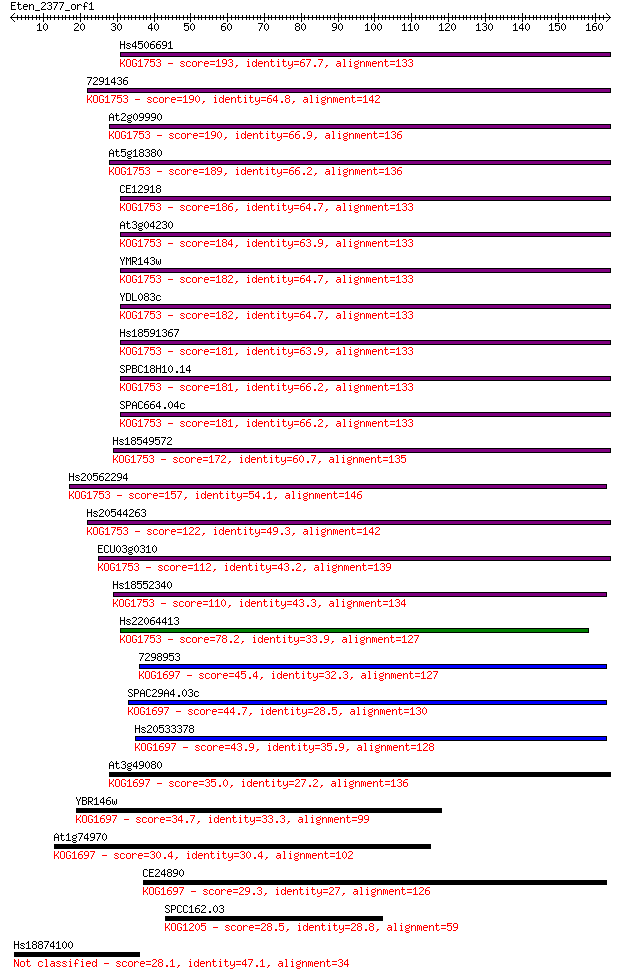
Query= Eten_2377_orf1
Length=163
Score E
Sequences producing significant alignments: (Bits) Value
Hs4506691 193 1e-49
7291436 190 9e-49
At2g09990 190 1e-48
At5g18380 189 2e-48
CE12918 186 2e-47
At3g04230 184 5e-47
YMR143w 182 3e-46
YDL083c 182 3e-46
Hs18591367 181 6e-46
SPBC18H10.14 181 8e-46
SPAC664.04c 181 8e-46
Hs18549572 172 3e-43
Hs20562294 157 7e-39
Hs20544263 122 3e-28
ECU03g0310 112 4e-25
Hs18552340 110 1e-24
Hs22064413 78.2 6e-15
7298953 45.4 5e-05
SPAC29A4.03c 44.7 8e-05
Hs20533378 43.9 1e-04
At3g49080 35.0 0.071
YBR146w 34.7 0.082
At1g74970 30.4 1.6
CE24890 29.3 3.5
SPCC162.03 28.5 5.9
Hs18874100 28.1 7.3
> Hs4506691
Length=146
Score = 193 bits (491), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 90/133 (67%), Positives = 111/133 (83%), Gaps = 0/133 (0%)
Query 31 VQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGIDIR 90
VQ FGRKKTA AVA C G GLI+VNG PL +++P L+ K E +L+LGK RF G+DIR
Sbjct 10 VQVFGRKKTATAVAHCKRGNGLIKVNGRPLEMIEPRTLQYKLLEPVLLLGKERFAGVDIR 69
Query 91 VRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRCEP 150
VRV+GGG V+Q+YAIRQ+I+K +VAY+QKYVDEA+K+EI+D L+ DR+L+VADPRRCE
Sbjct 70 VRVKGGGHVAQIYAIRQSISKALVAYYQKYVDEASKKEIKDILIQYDRTLLVADPRRCES 129
Query 151 KKFGGPGARARYQ 163
KKFGGPGARARYQ
Sbjct 130 KKFGGPGARARYQ 142
> 7291436
Length=148
Score = 190 bits (483), Expect = 9e-49, Method: Compositional matrix adjust.
Identities = 92/144 (63%), Positives = 115/144 (79%), Gaps = 2/144 (1%)
Query 22 MSEARR--TRRVQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVL 79
M + RR + VQ FGRKKTA AVA C G GL++VNG PL ++P+ L+ K E +L+L
Sbjct 1 MQQKRREPVQAVQVFGRKKTATAVAYCKRGNGLLKVNGRPLEQIEPKVLQYKLQEPLLLL 60
Query 80 GKNRFEGIDIRVRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRS 139
GK +F G+DIRVRV GGG V+Q+YAIRQAI+K +VA++QKYVDEA+K+EI+D LV DR+
Sbjct 61 GKEKFAGVDIRVRVSGGGHVAQIYAIRQAISKALVAFYQKYVDEASKKEIKDILVQYDRT 120
Query 140 LVVADPRRCEPKKFGGPGARARYQ 163
L+V DPRRCEPKKFGGPGARARYQ
Sbjct 121 LLVGDPRRCEPKKFGGPGARARYQ 144
> At2g09990
Length=146
Score = 190 bits (483), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 91/136 (66%), Positives = 110/136 (80%), Gaps = 0/136 (0%)
Query 28 TRRVQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGI 87
T VQ FGRKKTAVAV C G GLI++NG P+ L +PE LR K E IL+LGK+RF G+
Sbjct 7 TESVQCFGRKKTAVAVTHCKRGSGLIKLNGCPIELFQPEILRFKIFEPILLLGKHRFAGV 66
Query 88 DIRVRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRR 147
++R+RV GGG SQVYAIRQ+IAK +VAY+QKYVDE +K+EI+D LV DR+L+VADPRR
Sbjct 67 NMRIRVNGGGHTSQVYAIRQSIAKALVAYYQKYVDEQSKKEIKDILVRYDRTLLVADPRR 126
Query 148 CEPKKFGGPGARARYQ 163
CEPKKFGG GAR+RYQ
Sbjct 127 CEPKKFGGRGARSRYQ 142
> At5g18380
Length=146
Score = 189 bits (481), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 90/136 (66%), Positives = 110/136 (80%), Gaps = 0/136 (0%)
Query 28 TRRVQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGI 87
T VQ FGRKKTAVAV C G GLI++NG P+ L +PE LR K E +L+LGK+RF G+
Sbjct 7 TESVQCFGRKKTAVAVTHCKRGSGLIKLNGCPIELFQPEILRFKIFEPVLLLGKHRFAGV 66
Query 88 DIRVRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRR 147
++R+RV GGG SQVYAIRQ+IAK +VAY+QKYVDE +K+EI+D LV DR+L+VADPRR
Sbjct 67 NMRIRVNGGGHTSQVYAIRQSIAKALVAYYQKYVDEQSKKEIKDILVRYDRTLLVADPRR 126
Query 148 CEPKKFGGPGARARYQ 163
CEPKKFGG GAR+RYQ
Sbjct 127 CEPKKFGGRGARSRYQ 142
> CE12918
Length=144
Score = 186 bits (472), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 86/133 (64%), Positives = 110/133 (82%), Gaps = 0/133 (0%)
Query 31 VQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGIDIR 90
VQTFGRKKTA AVA C +G+GLI+VNG PL L+P+ LR+K E +L++GK RF+ +DIR
Sbjct 8 VQTFGRKKTATAVAHCKKGQGLIKVNGRPLEFLEPQILRIKLQEPLLLVGKERFQDVDIR 67
Query 91 VRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRCEP 150
+RV GGG V+Q+YA+RQA+AK +VAY+ KYVDE +KRE+++ A D+SL+VADPRR E
Sbjct 68 IRVSGGGHVAQIYAVRQALAKALVAYYHKYVDEQSKRELKNIFAAYDKSLLVADPRRRES 127
Query 151 KKFGGPGARARYQ 163
KKFGGPGARARYQ
Sbjct 128 KKFGGPGARARYQ 140
> At3g04230
Length=146
Score = 184 bits (468), Expect = 5e-47, Method: Compositional matrix adjust.
Identities = 85/133 (63%), Positives = 108/133 (81%), Gaps = 0/133 (0%)
Query 31 VQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGIDIR 90
VQ FGRKKTA AV C G G+I++NG+P+ L +PE LR K E +L+LGK+RF G+D+R
Sbjct 10 VQCFGRKKTATAVTYCKRGSGMIKLNGSPIELYQPEILRFKIFEPVLLLGKHRFAGVDMR 69
Query 91 VRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRCEP 150
+R GGG S+VYAIRQ+IAK +VAY+QKYVDE +K+EI+D L+ DR+L+VADPRRCE
Sbjct 70 IRATGGGNTSRVYAIRQSIAKALVAYYQKYVDEQSKKEIKDILMRYDRTLLVADPRRCES 129
Query 151 KKFGGPGARARYQ 163
KKFGGPGARAR+Q
Sbjct 130 KKFGGPGARARFQ 142
> YMR143w
Length=143
Score = 182 bits (461), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 86/133 (64%), Positives = 108/133 (81%), Gaps = 0/133 (0%)
Query 31 VQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGIDIR 90
VQTFG+KK+A AVA GKGLI+VNG+P+ L++PE LR K E +L++G ++F IDIR
Sbjct 7 VQTFGKKKSATAVAHVKAGKGLIKVNGSPITLVEPEILRFKVYEPLLLVGLDKFSNIDIR 66
Query 91 VRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRCEP 150
VRV GGG VSQVYAIRQAIAKG+VAYHQKYVDE +K E++ A + DR+L++AD RR EP
Sbjct 67 VRVTGGGHVSQVYAIRQAIAKGLVAYHQKYVDEQSKNELKKAFTSYDRTLLIADSRRPEP 126
Query 151 KKFGGPGARARYQ 163
KKFGG GAR+R+Q
Sbjct 127 KKFGGKGARSRFQ 139
> YDL083c
Length=143
Score = 182 bits (461), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 86/133 (64%), Positives = 108/133 (81%), Gaps = 0/133 (0%)
Query 31 VQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGIDIR 90
VQTFG+KK+A AVA GKGLI+VNG+P+ L++PE LR K E +L++G ++F IDIR
Sbjct 7 VQTFGKKKSATAVAHVKAGKGLIKVNGSPITLVEPEILRFKVYEPLLLVGLDKFSNIDIR 66
Query 91 VRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRCEP 150
VRV GGG VSQVYAIRQAIAKG+VAYHQKYVDE +K E++ A + DR+L++AD RR EP
Sbjct 67 VRVTGGGHVSQVYAIRQAIAKGLVAYHQKYVDEQSKNELKKAFTSYDRTLLIADSRRPEP 126
Query 151 KKFGGPGARARYQ 163
KKFGG GAR+R+Q
Sbjct 127 KKFGGKGARSRFQ 139
> Hs18591367
Length=146
Score = 181 bits (459), Expect = 6e-46, Method: Compositional matrix adjust.
Identities = 85/133 (63%), Positives = 107/133 (80%), Gaps = 0/133 (0%)
Query 31 VQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGIDIR 90
VQ FGRKKTA AVA C G GLI+VNG PL + +P L+ + E +L+LGK RF G+DIR
Sbjct 10 VQVFGRKKTATAVAHCRRGNGLIKVNGQPLEMTEPRMLQYELLEPVLLLGKERFAGVDIR 69
Query 91 VRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRCEP 150
VRV+GG V+Q+YAIRQ+ +K +VAY+QKYVDEA+K+EI+D L+ DR+L+VADPRRC+
Sbjct 70 VRVKGGAHVAQIYAIRQSTSKALVAYYQKYVDEASKKEIKDILIQYDRTLLVADPRRCKS 129
Query 151 KKFGGPGARARYQ 163
KKFGGPGARA YQ
Sbjct 130 KKFGGPGARACYQ 142
> SPBC18H10.14
Length=140
Score = 181 bits (458), Expect = 8e-46, Method: Compositional matrix adjust.
Identities = 88/133 (66%), Positives = 107/133 (80%), Gaps = 0/133 (0%)
Query 31 VQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGIDIR 90
VQ FG+K A AVA C GKGLI+VNGAPL L++PE LR+K E ILV G ++F G+DIR
Sbjct 4 VQCFGKKGNATAVAHCKVGKGLIKVNGAPLSLVQPEILRMKVYEPILVAGADKFAGVDIR 63
Query 91 VRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRCEP 150
VRV GGG VSQ+YAIRQAI+K +VAY+QK+VDE +K E++ AL+ DR+L+VADPRR EP
Sbjct 64 VRVSGGGHVSQIYAIRQAISKAIVAYYQKFVDEHSKAELKKALITYDRTLLVADPRRMEP 123
Query 151 KKFGGPGARARYQ 163
KKFGG GARAR Q
Sbjct 124 KKFGGHGARARQQ 136
> SPAC664.04c
Length=140
Score = 181 bits (458), Expect = 8e-46, Method: Compositional matrix adjust.
Identities = 88/133 (66%), Positives = 107/133 (80%), Gaps = 0/133 (0%)
Query 31 VQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGIDIR 90
VQ FG+K A AVA C GKGLI+VNGAPL L++PE LR+K E ILV G ++F G+DIR
Sbjct 4 VQCFGKKGNATAVAHCKVGKGLIKVNGAPLSLVQPEILRMKVYEPILVAGADKFAGVDIR 63
Query 91 VRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRCEP 150
VRV GGG VSQ+YAIRQAI+K +VAY+QK+VDE +K E++ AL+ DR+L+VADPRR EP
Sbjct 64 VRVSGGGHVSQIYAIRQAISKAIVAYYQKFVDEHSKAELKKALITYDRTLLVADPRRMEP 123
Query 151 KKFGGPGARARYQ 163
KKFGG GARAR Q
Sbjct 124 KKFGGHGARARQQ 136
> Hs18549572
Length=146
Score = 172 bits (435), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 82/135 (60%), Positives = 106/135 (78%), Gaps = 0/135 (0%)
Query 29 RRVQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGID 88
+ VQ FGRKKTA AVA C G GLI+VNG PL +++ L+ K E +L+LGK RF G+D
Sbjct 8 QSVQVFGRKKTATAVAHCKRGNGLIKVNGWPLEMIELRMLQYKLLEPVLLLGKERFAGVD 67
Query 89 IRVRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRC 148
I VR++GGG V+Q+YAI Q+I+K +VAY+QKYVDEA+K+EI+D L+ D++L+VADPR
Sbjct 68 ICVRMKGGGHVAQIYAIHQSISKALVAYYQKYVDEASKKEIKDILIQYDQTLLVADPRCR 127
Query 149 EPKKFGGPGARARYQ 163
E KKFGGPGA ARYQ
Sbjct 128 ESKKFGGPGAHARYQ 142
> Hs20562294
Length=496
Score = 157 bits (398), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 79/146 (54%), Positives = 104/146 (71%), Gaps = 0/146 (0%)
Query 17 DSIKKMSEARRTRRVQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESI 76
++ K + R + V+ FG KKTA AVA C G GLI+VNG PL ++P L+ K E +
Sbjct 351 NTCKPQIKKWRRQSVRVFGHKKTATAVAHCKRGNGLIKVNGRPLEKIEPPTLQYKLLEPV 410
Query 77 LVLGKNRFEGIDIRVRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVAC 136
L+L K RF G+DIRVRV GGG V+Q+YA RQ+I+K +VAY+QKY+DEA+K+EI+ L+
Sbjct 411 LLLSKERFAGVDIRVRVEGGGHVAQIYATRQSISKALVAYYQKYMDEASKKEIKGILILY 470
Query 137 DRSLVVADPRRCEPKKFGGPGARARY 162
D +L+VADP CE KKF PGA ARY
Sbjct 471 DWTLLVADPCPCESKKFRSPGACARY 496
> Hs20544263
Length=141
Score = 122 bits (306), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 70/144 (48%), Positives = 88/144 (61%), Gaps = 9/144 (6%)
Query 22 MSEARRTRRVQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLG- 80
M + + VQ FG KKTA AV C G GLI+VNG L ++K LR K E +L LG
Sbjct 1 MRSEGQLQSVQVFGGKKTATAVVHCKHGDGLIKVNGRLLEMIKLRTLRYKLLEPVLFLGP 60
Query 81 -KNRFEGIDIRVRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRS 139
K RF G+DI V V+G F+ V+ +QKY DE++K EI+D L+ D++
Sbjct 61 GKERFAGVDIHVHVKGPRFMLSVHLQSPG------GLYQKYADESSK-EIKDILIQYDQT 113
Query 140 LVVADPRRCEPKKFGGPGARARYQ 163
L+VADPRR E KKFGGPGARA YQ
Sbjct 114 LLVADPRRSESKKFGGPGARACYQ 137
> ECU03g0310
Length=145
Score = 112 bits (279), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 60/140 (42%), Positives = 87/140 (62%), Gaps = 1/140 (0%)
Query 25 ARRTRRVQTFGRKKTAVAVALCTE-GKGLIRVNGAPLHLLKPEALRVKACESILVLGKNR 83
+R + V T G+KK AVA +CT+ G+ IR+N P L+ + K E I ++ +
Sbjct 2 SRSVKSVTTQGKKKRAVATCVCTQTGEFSIRINKVPFKLITNTLMIAKLKEIICIVEQAN 61
Query 84 FEGIDIRVRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVA 143
+ + + + GG VS+VYA+R A AK ++A++ Y DE K+EI+ L+ DR +VA
Sbjct 62 IKDLSFDITCKIGGDVSRVYAVRMAFAKAILAFYGTYFDEWKKQEIQKRLLDFDRFSLVA 121
Query 144 DPRRCEPKKFGGPGARARYQ 163
D R+ EPKKFGGPGARARYQ
Sbjct 122 DSRKREPKKFGGPGARARYQ 141
> Hs18552340
Length=132
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 58/134 (43%), Positives = 84/134 (62%), Gaps = 9/134 (6%)
Query 29 RRVQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGID 88
+ Q FG KK A AVA GLI+VNG L +++ L+ K E +L+ K+
Sbjct 8 QYAQVFGCKKRATAVAPGKHSNGLIKVNGWHLEMIELHMLQYKLLEPLLLWAKSD----- 62
Query 89 IRVRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRC 148
+ GG V+Q+ A+ Q+I+K +VAY+QKY+DEA+K++I+D L+ D++L+VA RC
Sbjct 63 ----LLGGSHVAQICAVHQSISKALVAYYQKYMDEASKKDIKDTLIQNDQTLLVAGLLRC 118
Query 149 EPKKFGGPGARARY 162
E KKFG GA ARY
Sbjct 119 ESKKFGSSGACARY 132
> Hs22064413
Length=248
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 43/134 (32%), Positives = 68/134 (50%), Gaps = 27/134 (20%)
Query 31 VQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGIDIR 90
+Q G++K A+A G GL+ VN PL +++ L K E +L+LGK RF +DI
Sbjct 10 MQVSGQRKMTTAMAGRKHGSGLMEVNRRPLEMMESCTLHYKLLEPVLLLGKERFADVDIH 69
Query 91 VRVRG-------GGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVA 143
V ++G GG + ++ ++++D L+ C +L+VA
Sbjct 70 VLLKGDVHLQSPGGLLPEI--------------------RVASKKVKDVLIQCGWTLLVA 109
Query 144 DPRRCEPKKFGGPG 157
DP RC+ +KFGGPG
Sbjct 110 DPHRCKSRKFGGPG 123
> 7298953
Length=395
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 41/130 (31%), Positives = 57/130 (43%), Gaps = 18/130 (13%)
Query 36 RKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESIL---VLGKNRFEGIDIRVR 92
RK V + G G I +NG + + E R + + +LGK +D+
Sbjct 276 RKTARADVTVRLPGTGKISINGKDISYFEDENCREQLLFPLQFSELLGK-----VDVEAN 330
Query 93 VRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRCEPKK 152
V GGG Q AIR IA + + +VD+ +R A L+ D RR E KK
Sbjct 331 VEGGGPSGQAGAIRWGIAMSL----RSFVDQEMIESMRLA------GLLTRDYRRRERKK 380
Query 153 FGGPGARARY 162
FG GAR +Y
Sbjct 381 FGQEGARRKY 390
> SPAC29A4.03c
Length=132
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 57/130 (43%), Gaps = 12/130 (9%)
Query 33 TFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGIDIRVR 92
T G++K++ A G G VNG+P + + K L NR ++
Sbjct 10 TKGKRKSSKATVKMLPGTGKFYVNGSPFDVYFQRMVHRKHAVYPLA-ACNRLTNYNVWAT 68
Query 93 VRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRCEPKK 152
V GGG Q A+ AI+K ++ + E + ++ V D V+ D R+ E KK
Sbjct 69 VHGGGPTGQSGAVHAAISKSLI------LQEPSLKQ-----VIKDTHCVLNDKRKVERKK 117
Query 153 FGGPGARARY 162
G P AR +Y
Sbjct 118 TGQPKARKKY 127
> Hs20533378
Length=396
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 46/134 (34%), Positives = 62/134 (46%), Gaps = 22/134 (16%)
Query 35 GRKKTAVAVALC-TEGKGLIRVNGAPLHLLKP-----EALRVKACESILVLGKNRFEGID 88
G++KTA A A+ G G I+VNG L P E L + LGK+ D
Sbjct 274 GKRKTAKAEAIVYKHGSGRIKVNGIDYQLYFPITQDREQLMF-PFHFVDRLGKH-----D 327
Query 89 IRVRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVADPRRC 148
+ V GGG +Q AIR A+AK + + +V E +R A L+ DPR
Sbjct 328 VTCTVSGGGRSAQAGAIRLAMAKALCS----FVTEDEVEWMRQA------GLLTTDPRVR 377
Query 149 EPKKFGGPGARARY 162
E KK G GAR ++
Sbjct 378 ERKKPGQEGARRKF 391
> At3g49080
Length=143
Score = 35.0 bits (79), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 37/140 (26%), Positives = 58/140 (41%), Gaps = 20/140 (14%)
Query 28 TRRVQTFGRKKTAVAVALCTEGKGLIRVNGAPLH----LLKPEALRVKACESILVLGKNR 83
T R GR+K ++A G+G +VN +L A ++ LG+
Sbjct 16 TGRAYGTGRRKCSIARVWIQPGEGKFQVNEKEFDVYFPMLDHRAALLRPLAETKTLGR-- 73
Query 84 FEGIDIRVRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLVVA 143
DI+ V+GGG QV AI+ I++ + + ++R +L A +
Sbjct 74 ---WDIKCTVKGGGTTGQVGAIQLGISRALQNWEP---------DMRTSLRAA--GFLTR 119
Query 144 DPRRCEPKKFGGPGARARYQ 163
D R E KK G AR +Q
Sbjct 120 DSRVVERKKPGKAKARKSFQ 139
> YBR146w
Length=278
Score = 34.7 bits (78), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 49/106 (46%), Gaps = 18/106 (16%)
Query 19 IKKMSEARRTRRVQTFGRKKTAVAVALCTEGKGLIRVNGAPLHLLKPEALRVKACESILV 78
IK + E R+ V G++K++ A G G I VNG L+ L++K ESI+
Sbjct 145 IKTLDEFGRSIAV---GKRKSSTAKVFVVRGTGEILVNGRQLN---DYFLKMKDRESIMY 198
Query 79 -------LGKNRFEGIDIRVRVRGGGFVSQVYAIRQAIAKGVVAYH 117
+GK +I GGG Q +I AIAK +V ++
Sbjct 199 PLQVIESVGK-----YNIFATTSGGGPTGQAESIMHAIAKALVVFN 239
> At1g74970
Length=208
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 31/120 (25%), Positives = 56/120 (46%), Gaps = 28/120 (23%)
Query 13 PPSPDSIKKMSEARRTR--------RVQTFGRKKTAVAVALCTEGKGLIRVN-------- 56
PP + I ++ + ++R ++ GR+K A+A + EG G + +N
Sbjct 57 PPEEEEIVELKKYVKSRLPGGFAAQKIIGTGRRKCAIARVVLQEGTGKVIINYRDAKEYL 116
Query 57 -GAPLHLLKPEALRVKACESILVLG-KNRFEGIDIRVRVRGGGFVSQVYAIRQAIAKGVV 114
G PL L + ++V ++ LG +N + DI V+ GGG Q AI +A+ ++
Sbjct 117 QGNPLWL---QYVKV----PLVTLGYENSY---DIFVKAHGGGLSGQAQAITLGVARALL 166
> CE24890
Length=392
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 34/141 (24%), Positives = 53/141 (37%), Gaps = 26/141 (18%)
Query 37 KKTAVAVALCTEGKGLIRVNGAPLH----------LLKPEAL-----RVKACESILVLGK 81
K T V + GKG ++G LH LL P + R + +
Sbjct 258 KDTRATVKVTDAGKGKFDIDGLQLHDFRHLQAREILLAPMIVSQSLGRFDVTATTSCISN 317
Query 82 NRFEGIDIRVRVRGGGFVSQVYAIRQAIAKGVVAYHQKYVDEATKREIRDALVACDRSLV 141
E + +R GG + A+R A V A + ++ +R + L+
Sbjct 318 TLPEAPNKAPLMRSGGMSALPRAVRHGTALCVAALQPEAIEP-----LRLS------GLL 366
Query 142 VADPRRCEPKKFGGPGARARY 162
DPR+ E K PGARA++
Sbjct 367 TLDPRKNERSKVNQPGARAKW 387
> SPCC162.03
Length=292
Score = 28.5 bits (62), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 26/59 (44%), Gaps = 0/59 (0%)
Query 43 VALCTEGKGLIRVNGAPLHLLKPEALRVKACESILVLGKNRFEGIDIRVRVRGGGFVSQ 101
V C+ I + + L LK + VK+ E+ K RF +DI + G G V +
Sbjct 32 VIACSRAPDTITIEHSKLLKLKLDVTDVKSVETAFKDAKRRFGNVDIVINNAGYGLVGE 90
> Hs18874100
Length=569
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 20/42 (47%), Gaps = 8/42 (19%)
Query 2 EGNPPLLSFCLPPSPDSIKKMSEARR--------TRRVQTFG 35
+G P S CLPP PDS+ S ++R T RVQ
Sbjct 521 DGPPDSTSTCLPPEPDSLLGCSSSQRAASLDSVATSRVQDLS 562
Lambda K H
0.322 0.139 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2281134618
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40