bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2349_orf1
Length=599
Score E
Sequences producing significant alignments: (Bits) Value
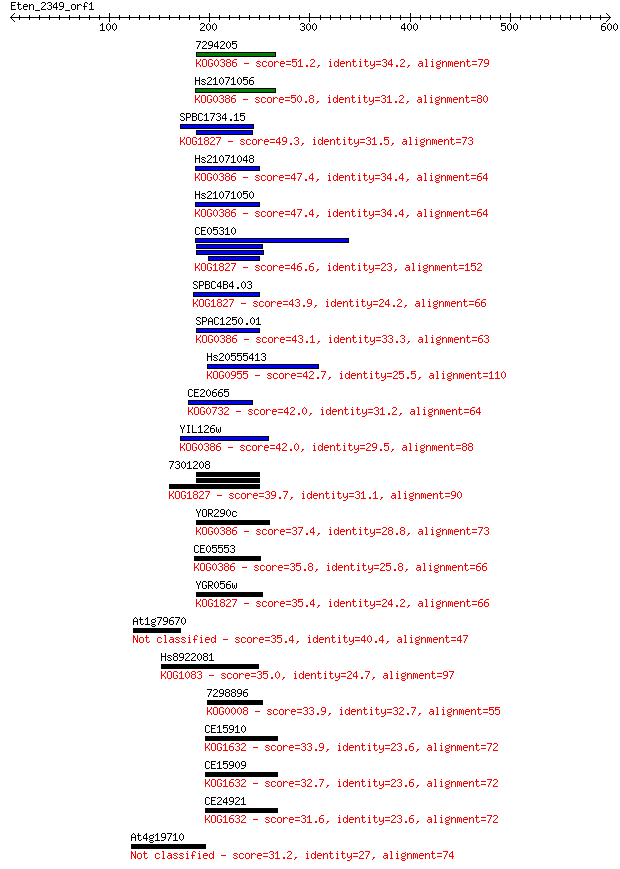
7294205 51.2 6e-06
Hs21071056 50.8 8e-06
SPBC1734.15 49.3 3e-05
Hs21071048 47.4 1e-04
Hs21071050 47.4 1e-04
CE05310 46.6 2e-04
SPBC4B4.03 43.9 0.001
SPAC1250.01 43.1 0.002
Hs20555413 42.7 0.002
CE20665 42.0 0.004
YIL126w 42.0 0.004
7301208 39.7 0.018
YOR290c 37.4 0.11
CE05553 35.8 0.27
YGR056w 35.4 0.34
At1g79670 35.4 0.36
Hs8922081 35.0 0.52
7298896 33.9 1.1
CE15910 33.9 1.2
CE15909 32.7 2.7
CE24921 31.6 5.3
At4g19710 31.2 6.2
> 7294205
Length=1638
Score = 51.2 bits (121), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 47/80 (58%), Gaps = 1/80 (1%)
Query 187 FVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQES 246
F++LPS DYYE + +PV + +I + + ++ LN++EK +L +NA+ YN + S
Sbjct 1451 FMKLPSRQRLPDYYEIIKRPVDIKKILQRIEDCKYADLNELEKDFMQLCQNAQIYNEEAS 1510
Query 247 PIFLRALECMG-FVLIRSRV 265
I+L ++ FV R R+
Sbjct 1511 LIYLDSIALQKVFVGARQRI 1530
> Hs21071056
Length=1647
Score = 50.8 bits (120), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 45/81 (55%), Gaps = 1/81 (1%)
Query 186 AFVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQE 245
F++LPS +YYE + KPV +I+ H++ SLN +EK + L +NA+ +N +
Sbjct 1484 VFIQLPSRKELPEYYELIRKPVDFKKIKERIRNHKYRSLNDLEKDVMLLCQNAQTFNLEG 1543
Query 246 SPIFLRALECMG-FVLIRSRV 265
S I+ ++ F +R ++
Sbjct 1544 SLIYEDSIVLQSVFTSVRQKI 1564
> SPBC1734.15
Length=542
Score = 49.3 bits (116), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 41/80 (51%), Gaps = 7/80 (8%)
Query 171 ISQALTEAIQESR-------FSAFVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTS 223
+ Q + +A+ E + F++LPS +Y DYYE + P+ + + + K E+T+
Sbjct 147 VIQNILDALHEEKDEQGRFLIDIFIDLPSKRLYPDYYEIIKSPMTIKMLEKRFKKGEYTT 206
Query 224 LNKVEKYLTRLTENARAYNG 243
L K L ++ NA+ YN
Sbjct 207 LESFVKDLNQMFINAKTYNA 226
Score = 40.4 bits (93), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 187 FVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYN 242
F ELPS + DYY+ + KP+C +R A ++ S+ + + NA+ YN
Sbjct 37 FEELPSKRYFPDYYQIIQKPICYKMMRNKAKTGKYLSMGDFYDDIRLMVSNAQTYN 92
> Hs21071048
Length=1586
Score = 47.4 bits (111), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 36/64 (56%), Gaps = 0/64 (0%)
Query 186 AFVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQE 245
F++LPS +YYE + KPV +I+ H++ SL +EK + L NA+ +N +
Sbjct 1422 VFIQLPSRKELPEYYELIRKPVDFKKIKERIRNHKYRSLGDLEKDVMLLCHNAQTFNLEG 1481
Query 246 SPIF 249
S I+
Sbjct 1482 SQIY 1485
> Hs21071050
Length=1568
Score = 47.4 bits (111), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 36/64 (56%), Gaps = 0/64 (0%)
Query 186 AFVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQE 245
F++LPS +YYE + KPV +I+ H++ SL +EK + L NA+ +N +
Sbjct 1404 VFIQLPSRKELPEYYELIRKPVDFKKIKERIRNHKYRSLGDLEKDVMLLCHNAQTFNLEG 1463
Query 246 SPIF 249
S I+
Sbjct 1464 SQIY 1467
> CE05310
Length=1879
Score = 46.6 bits (109), Expect = 2e-04, Method: Composition-based stats.
Identities = 35/152 (23%), Positives = 62/152 (40%), Gaps = 16/152 (10%)
Query 186 AFVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQE 245
AF++LPS Y +YY+ + P+ + IR D H++ ++ + + NAR +N
Sbjct 559 AFIQLPSAKQYPEYYQIIQNPIDMKTIRMRIDGHQYPQVDAMINDCRVMFSNARDFNEPR 618
Query 246 SPIFLRALECMGFVLIRSRVNACVAFHSMQSAAQGERVRRLFEVYRSCWDSTASTSLPDG 305
S I + A++ VL A+ M++ + G + S P
Sbjct 619 SMIHMDAIQLEKAVL--------RAYEGMRNTSLGSSIPSTPHSSSSNLMKAMKLKTPKS 670
Query 306 EASGAGGDGLGGELMEAEGEGDDDEASAVSSQ 337
E G G G DD+E++ ++ Q
Sbjct 671 EVRGRGKKNFSGS--------DDEESARINIQ 694
Score = 39.3 bits (90), Expect = 0.028, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query 187 FVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQES 246
F+ELPS Y DYY+ + PV + I + ++ L + L ++ NA YN + S
Sbjct 368 FLELPSKESYPDYYDEIKNPVSIFMINKRLKNGKY-DLKSLVADLMQMYSNAFDYNLESS 426
Query 247 PIFLRA 252
+++ A
Sbjct 427 EVYISA 432
Score = 35.0 bits (79), Expect = 0.44, Method: Composition-based stats.
Identities = 17/67 (25%), Positives = 35/67 (52%), Gaps = 0/67 (0%)
Query 187 FVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQES 246
F+ LPS + YY+ + KP+ +++I+ + ++ +L V + NA +N +S
Sbjct 737 FMRLPSKEEFPAYYDVIKKPMDMMRIKHKLENRQYVTLLDVVSDFMLMLSNACKFNETDS 796
Query 247 PIFLRAL 253
I+ A+
Sbjct 797 DIYKEAV 803
Score = 34.7 bits (78), Expect = 0.63, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 32/51 (62%), Gaps = 0/51 (0%)
Query 199 YYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQESPIF 249
YYE++AKP+ L I + +++++ +++ L L +NA+ ++G S IF
Sbjct 230 YYEKIAKPIDLKTIAQNGVNKKYSTMKELKDDLFLLFKNAQQFSGNGSDIF 280
> SPBC4B4.03
Length=803
Score = 43.9 bits (102), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 16/66 (24%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 184 FSAFVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNG 243
F+ F LP P ++ +YY+ + +P+ L I++ KH + ++ + + +NA+++N
Sbjct 234 FAPFERLPDPRMFPEYYQAIEQPMALEVIQKKLSKHRYETIEQFVDDFNLMFDNAKSFND 293
Query 244 QESPIF 249
S ++
Sbjct 294 PSSQVY 299
> SPAC1250.01
Length=1199
Score = 43.1 bits (100), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 0/63 (0%)
Query 187 FVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQES 246
F+ELPS +Y DYY + P+ L IR+ + + +L ++ L + NAR YN + S
Sbjct 1092 FLELPSKKLYPDYYMIIKSPIALDAIRKHINGTFYKTLEAMKSDLMTMFNNARTYNEEGS 1151
Query 247 PIF 249
++
Sbjct 1152 FVY 1154
> Hs20555413
Length=1205
Score = 42.7 bits (99), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/114 (24%), Positives = 53/114 (46%), Gaps = 5/114 (4%)
Query 198 DYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQESPIFLRA---LE 254
DY E ++KP+ +RR + H + +L + E+ + N YN +++ IF RA L
Sbjct 625 DYLEFISKPMDFSTMRRKLESHLYRTLEEFEEDFNLIVTNCMKYNAKDT-IFHRAAVRLR 683
Query 255 CMGFVLIR-SRVNACVAFHSMQSAAQGERVRRLFEVYRSCWDSTASTSLPDGEA 307
+G ++R +R A + + +L + YR W+ + +P+ A
Sbjct 684 DLGGAILRHARRQAENIGYDPERGTHLPESPKLEDFYRFSWEDVDNILIPENRA 737
> CE20665
Length=1242
Score = 42.0 bits (97), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 0/64 (0%)
Query 179 IQESRFSAFVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENA 238
+++ RF FVE P +DYYE + P+C+ I + E+ +K L + NA
Sbjct 882 MRDRRFVEFVEPVDPDEAEDYYEIIETPICMQDIMEKLNNCEYNHADKFVADLILIQTNA 941
Query 239 RAYN 242
YN
Sbjct 942 LEYN 945
> YIL126w
Length=1359
Score = 42.0 bits (97), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 26/88 (29%), Positives = 45/88 (51%), Gaps = 0/88 (0%)
Query 171 ISQALTEAIQESRFSAFVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKY 230
+ + L E R S F +LPS Y DY++ + KP+ + I + + +L +V +
Sbjct 1262 MREQLDEVDSHPRTSIFEKLPSKRDYPDYFKVIEKPMAIDIILKNCKNGTYKTLEEVRQA 1321
Query 231 LTRLTENARAYNGQESPIFLRALECMGF 258
L + ENAR YN + S +++ A + F
Sbjct 1322 LQTMFENARFYNEEGSWVYVDADKLNEF 1349
> 7301208
Length=1654
Score = 39.7 bits (91), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 36/63 (57%), Gaps = 0/63 (0%)
Query 187 FVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQES 246
F LPS +Y DYY+ + P+ L I + ++SL ++E+ L ++T+NA +N S
Sbjct 218 FQLLPSKKIYPDYYDVIEHPIDLRLIATKIQMNAYSSLAEMERDLLQMTKNACLFNEPGS 277
Query 247 PIF 249
I+
Sbjct 278 QIY 280
Score = 36.6 bits (83), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 18/63 (28%), Positives = 35/63 (55%), Gaps = 0/63 (0%)
Query 187 FVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQES 246
F +LPS + Y DYY+ + +P+ + +I + + + +L+ + + ENA YN +S
Sbjct 668 FTKLPSKSEYPDYYDIIREPIDMDRIAQKLKQGAYDTLDDLAADFLLMLENACKYNEPDS 727
Query 247 PIF 249
I+
Sbjct 728 QIY 730
Score = 33.1 bits (74), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 23/100 (23%), Positives = 41/100 (41%), Gaps = 11/100 (11%)
Query 160 PQRWRALLNGAISQALTEAIQESR----------FSAFVELPSPTVYKDYYERVAKPVCL 209
P+ R +N AI + + +IQ+ F+E P +Y DYY+ + P+ +
Sbjct 501 PKSNRIAINAAIKKKIL-SIQKYLVDYSLGNRRPIEMFMEKPPRKIYPDYYDIIQNPIDM 559
Query 210 LQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQESPIF 249
I + ++ V + N R YN + S I+
Sbjct 560 NTIEHNIRTDRYAAVEDVVSDYRLMFSNCRQYNEEGSNIY 599
> YOR290c
Length=1703
Score = 37.4 bits (85), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 33/73 (45%), Gaps = 0/73 (0%)
Query 187 FVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQES 246
F+ PS +Y DYY + PV I + + SL + + + NAR YN + S
Sbjct 1576 FLSKPSKALYPDYYMIIKYPVAFDNINTHIETLAYNSLKETLQDFHLIFSNARIYNTEGS 1635
Query 247 PIFLRALECMGFV 259
++ +LE V
Sbjct 1636 VVYEDSLELEKVV 1648
> CE05553
Length=1474
Score = 35.8 bits (81), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 34/66 (51%), Gaps = 0/66 (0%)
Query 185 SAFVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQ 244
F LP+ DYY+ ++KP+ +I + + +T + ++ + L NA+ YN +
Sbjct 1206 DVFQTLPTRKELPDYYQVISKPMDFDRINKKIETGRYTVMEELNDDMNLLVNNAQTYNEE 1265
Query 245 ESPIFL 250
S I++
Sbjct 1266 GSEIYV 1271
> YGR056w
Length=928
Score = 35.4 bits (80), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 16/66 (24%), Positives = 30/66 (45%), Gaps = 0/66 (0%)
Query 187 FVELPSPTVYKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQES 246
F +LP YY + P+CL+ IR+ ++ + + E+ + N + Y Q+
Sbjct 263 FEKLPDRNEEPTYYSVITDPICLMDIRKKVKSRKYRNFHTFEEDFQLMLTNFKLYYSQDQ 322
Query 247 PIFLRA 252
+RA
Sbjct 323 SNIIRA 328
> At1g79670
Length=751
Score = 35.4 bits (80), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 24/47 (51%), Gaps = 1/47 (2%)
Query 124 GVRGIPGGSSGCVDLWGCGEGQEGDSAGAAAACLADPQRWRALLNGA 170
G RG P GC+D+ C EG+ G S+ C+ P WR LNG
Sbjct 296 GYRGNPYLPGGCIDIDECEEGK-GLSSCGELTCVNVPGSWRCELNGV 341
> Hs8922081
Length=2969
Score = 35.0 bits (79), Expect = 0.52, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 51/97 (52%), Gaps = 4/97 (4%)
Query 152 AAAACLADPQRWRALLNGAISQALTEAIQESRFSAFVELPSPTVYKDYYERVAKPVCLLQ 211
A AA LA Q ++ + +G IS ++ +++ + + LP DYYE+++ P+ L+
Sbjct 2440 ARAARLA--QIFKEICDGIIS--YKDSSRQALAAPLLNLPPKKKNADYYEKISDPLDLIT 2495
Query 212 IRRFADKHEFTSLNKVEKYLTRLTENARAYNGQESPI 248
I + + ++ + + ++ NA Y G++SP+
Sbjct 2496 IEKQILTGYYKTVEAFDADMLKVFRNAEKYYGRKSPV 2532
> 7298896
Length=2065
Score = 33.9 bits (76), Expect = 1.1, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 198 DYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQESPIFLRA 252
DYY V KP+ L +R + + +TS + L ++ +N+ YNG +S L A
Sbjct 1506 DYYRVVTKPMDLQTMREYIRQRRYTSREMFLEDLKQIVDNSLIYNGPQSAYTLAA 1560
> CE15910
Length=510
Score = 33.9 bits (76), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 17/72 (23%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query 196 YKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQESPIFLRALEC 255
+ DY + + KP+ L I + ++ E+ L++ + ++ ENA+ YN + + +F + E
Sbjct 380 FPDYEKFIKKPMDLSTITKKVERTEYLYLSQFVNDVNQMFENAKTYNPKGNAVF-KCAET 438
Query 256 MGFVLIRSRVNA 267
M V + ++
Sbjct 439 MQEVFDKKLIDV 450
> CE15909
Length=405
Score = 32.7 bits (73), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 17/72 (23%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query 196 YKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQESPIFLRALEC 255
+ DY + + KP+ L I + ++ E+ L++ + ++ ENA+ YN + + +F + E
Sbjct 275 FPDYEKFIKKPMDLSTITKKVERTEYLYLSQFVNDVNQMFENAKTYNPKGNAVF-KCAET 333
Query 256 MGFVLIRSRVNA 267
M V + ++
Sbjct 334 MQEVFDKKLIDV 345
> CE24921
Length=451
Score = 31.6 bits (70), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 17/72 (23%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query 196 YKDYYERVAKPVCLLQIRRFADKHEFTSLNKVEKYLTRLTENARAYNGQESPIFLRALEC 255
+ DY + + KP+ L I + ++ E+ L++ + ++ ENA+ YN + + +F + E
Sbjct 321 FPDYEKFIKKPMDLSTITKKVERTEYLYLSQFVNDVNQMFENAKTYNPKGNAVF-KCAET 379
Query 256 MGFVLIRSRVNA 267
M V + ++
Sbjct 380 MQEVFDKKLIDV 391
> At4g19710
Length=916
Score = 31.2 bits (69), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 20/77 (25%), Positives = 37/77 (48%), Gaps = 3/77 (3%)
Query 122 GPGVRGIPGGSS---GCVDLWGCGEGQEGDSAGAAAACLADPQRWRALLNGAISQALTEA 178
G G+ G+PG +S GCV G ++ + C A P++ ++ A+ +EA
Sbjct 414 GTGMAGVPGTASDIFGCVKDVGANVIMISQASSEHSVCFAVPEKEVNAVSEALRSRFSEA 473
Query 179 IQESRFSAFVELPSPTV 195
+Q R S +P+ ++
Sbjct 474 LQAGRLSQIEVIPNCSI 490
Lambda K H
0.311 0.126 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 16794388910
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40