bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2337_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
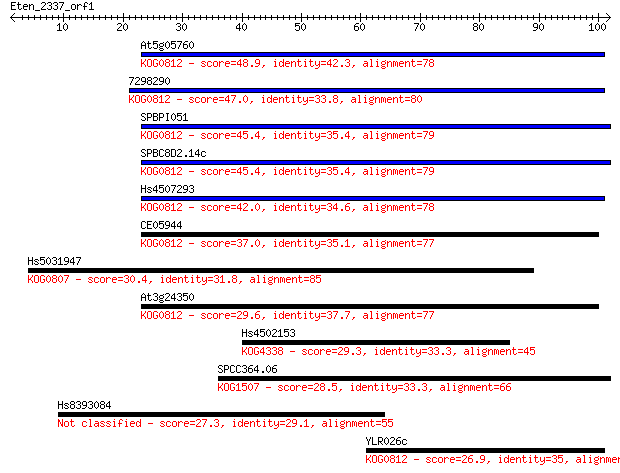
At5g05760 48.9 2e-06
7298290 47.0 1e-05
SPBPI051 45.4 3e-05
SPBC8D2.14c 45.4 3e-05
Hs4507293 42.0 3e-04
CE05944 37.0 0.010
Hs5031947 30.4 0.75
At3g24350 29.6 1.6
Hs4502153 29.3 2.0
SPCC364.06 28.5 2.8
Hs8393084 27.3 6.5
YLR026c 26.9 9.9
> At5g05760
Length=336
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 45/87 (51%), Gaps = 12/87 (13%)
Query 23 DRTAEFAAAAESFN-FAASP--------PAADLRQPFSDADKNFNLIASDMGNSLHATSL 73
DRT E + +++ A P PA+ R S FN AS +G + TS
Sbjct 7 DRTVELHSLSQTLKKIGAIPSVHQDEDDPASSKR---SSPGSEFNKKASRIGLGIKETSQ 63
Query 74 KLQELAKLARQCGIYNDKTAQIQELTF 100
K+ LAKLA+Q I+ND+T +IQELT
Sbjct 64 KITRLAKLAKQSTIFNDRTVEIQELTV 90
> 7298290
Length=310
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 46/84 (54%), Gaps = 4/84 (4%)
Query 21 AYDRTAEFAAAAESFNFAASPPAADLRQPFS----DADKNFNLIASDMGNSLHATSLKLQ 76
A DRT EFA A S A ++R P + F ++A +G ++ +T KL+
Sbjct 3 ARDRTGEFANAIRSLQARNITRAVNIRDPRKAKQVQSYSEFMMVARFIGKNIASTYAKLE 62
Query 77 ELAKLARQCGIYNDKTAQIQELTF 100
+L LA++ +++D+ +IQELT+
Sbjct 63 KLTMLAKKKSLFDDRPQEIQELTY 86
> SPBPI051
Length=309
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 44/83 (53%), Gaps = 4/83 (4%)
Query 23 DRTAEFAAAAESFNFAASPPAADLRQPFSDADKN----FNLIASDMGNSLHATSLKLQEL 78
DRTAEF A A+ D K+ F IA + N ++ T KLQ+L
Sbjct 5 DRTAEFQACVTKTRSRLRTTTANQAVGGPDQTKHQKSEFTRIAQKIANQINQTGEKLQKL 64
Query 79 AKLARQCGIYNDKTAQIQELTFE 101
++LA++ +++D+ +IQELTF+
Sbjct 65 SQLAKRKTLFDDRPVEIQELTFQ 87
> SPBC8D2.14c
Length=309
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 44/83 (53%), Gaps = 4/83 (4%)
Query 23 DRTAEFAAAAESFNFAASPPAADLRQPFSDADKN----FNLIASDMGNSLHATSLKLQEL 78
DRTAEF A A+ D K+ F IA + N ++ T KLQ+L
Sbjct 5 DRTAEFQACVTKTRSRLRTTTANQAVGGPDQTKHQKSEFTRIAQKIANQINQTGEKLQKL 64
Query 79 AKLARQCGIYNDKTAQIQELTFE 101
++LA++ +++D+ +IQELTF+
Sbjct 65 SQLAKRKTLFDDRPVEIQELTFQ 87
> Hs4507293
Length=301
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 44/83 (53%), Gaps = 9/83 (10%)
Query 23 DRTAEFAAAAESF-----NFAASPPAADLRQPFSDADKNFNLIASDMGNSLHATSLKLQE 77
DRT EF +A +S + PA + S+ F L+A +G L T KL++
Sbjct 5 DRTQEFLSACKSLQTRQNGIQTNKPALRAVRQRSE----FTLMAKRIGKDLSNTFAKLEK 60
Query 78 LAKLARQCGIYNDKTAQIQELTF 100
L LA++ +++DK +I+ELT+
Sbjct 61 LTILAKRKSLFDDKAVEIEELTY 83
> CE05944
Length=413
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 45/81 (55%), Gaps = 7/81 (8%)
Query 23 DRTAEFAAAAESFNFAASPPAADLR-QPFSDA---DKNFNLIASDMGNSLHATSLKLQEL 78
DRT+EF A A+S+ A+ A +R QP + FN +A +G L T K+++L
Sbjct 111 DRTSEFRATAKSYEMKAA--ANGIRPQPKHEMLSESVQFNQLAKRIGKELSQTCAKMEKL 168
Query 79 AKLARQCGIYNDKTAQIQELT 99
A+ A++ Y +++ QI L+
Sbjct 169 AEYAKKKSCYEERS-QIDHLS 188
> Hs5031947
Length=327
Score = 30.4 bits (67), Expect = 0.75, Method: Composition-based stats.
Identities = 27/89 (30%), Positives = 38/89 (42%), Gaps = 9/89 (10%)
Query 4 SSSSSSSSSSSSSQPTMAYDRTAEFAAAAESFNFAASPPAADLRQPFSDADKNFNLIASD 63
SSSS + Q T D+ F AE AA A P + F+ IA D
Sbjct 40 SSSSCELPLVAVCQVTSTPDKQQNFKTCAELVREAARLGACLAFLP-----EAFDFIARD 94
Query 64 MGNSLHATS----LKLQELAKLARQCGIY 88
+LH + L+E +LAR+CG++
Sbjct 95 PAETLHLSEPLGGKLLEEYTQLARECGLW 123
> At3g24350
Length=377
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 29/120 (24%), Positives = 49/120 (40%), Gaps = 44/120 (36%)
Query 23 DRTAEFAAAAESFNFAASPPAADLRQPFSDADKN-------------FNLIASDMGNSLH 69
DR+ EF E+ + +P A P+ + ++N FN AS +G +++
Sbjct 12 DRSDEFFKIVETLRRSIAPAPAANNVPYGN-NRNDGARREDLINKSEFNKRASHIGLAIN 70
Query 70 ATSLKLQELAK------------------------------LARQCGIYNDKTAQIQELT 99
TS KL +LAK +A++ +++D T +IQELT
Sbjct 71 QTSQKLSKLAKRIRMVLRSRTDLFSGVNFSSAYLSTRECVIVAKRTSVFDDPTQEIQELT 130
> Hs4502153
Length=4563
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 28/50 (56%), Gaps = 5/50 (10%)
Query 40 SPPAADLRQ-----PFSDADKNFNLIASDMGNSLHATSLKLQELAKLARQ 84
SP AD+ + P+ ++ N +AS + N L++ L +Q+L KL ++
Sbjct 568 SPSQADINKIVQILPWEQNEQVKNFVASHIANILNSEELDIQDLKKLVKE 617
> SPCC364.06
Length=393
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 33/74 (44%), Gaps = 11/74 (14%)
Query 36 NFAASPPAADLRQPFSDADKNFNLIASDMGNSLHATSLKLQELAKLA--RQCGI------ 87
NF A P + + D+N I +G+ LH TS + EL + R G+
Sbjct 32 NFGAKAPQVNT---IDEGDENLEAIDGKLGSLLHLTSEGVSELPEAVQRRISGLRGLQKR 88
Query 88 YNDKTAQIQELTFE 101
Y+D +Q Q+ FE
Sbjct 89 YSDLESQFQKELFE 102
> Hs8393084
Length=1240
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 28/57 (49%), Gaps = 2/57 (3%)
Query 9 SSSSSSSSQPTMAYDRTAEFAAAAESFNFAASPPAADLRQPF--SDADKNFNLIASD 63
+ S++ QP M RT++ A+ A+SPP + P A+ NF ++ +D
Sbjct 633 ARSAAGEGQPAMITFRTSKEKTASSKNTQASSPPVGIPKYPVVSEAANNNFGVVLTD 689
> YLR026c
Length=340
Score = 26.9 bits (58), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 61 ASDMGNSLHATSLKLQELAKLARQCGIYNDKTAQIQELTF 100
AS + + + +T+ L +LA LA++ ++ND +I EL+F
Sbjct 58 ASGIAHEISSTAQLLSKLAVLAKRKPMFNDNPVEIAELSF 97
Lambda K H
0.307 0.116 0.303
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184494980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40