bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
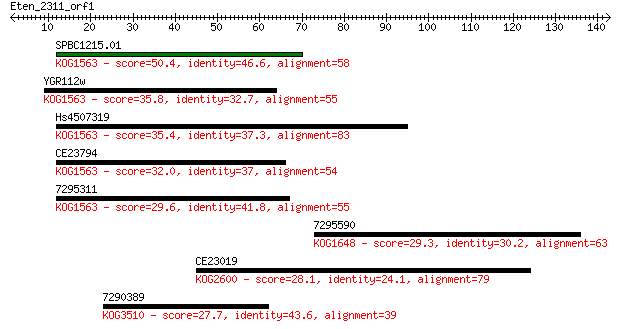
Query= Eten_2311_orf1
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
SPBC1215.01 50.4 1e-06
YGR112w 35.8 0.027
Hs4507319 35.4 0.036
CE23794 32.0 0.36
7295311 29.6 1.8
7295590 29.3 2.7
CE23019 28.1 5.7
7290389 27.7 7.2
> SPBC1215.01
Length=290
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/58 (46%), Positives = 34/58 (58%), Gaps = 2/58 (3%)
Query 12 YTPVVLHGVLDSSTELLVGPRPGLDVGNPGYCVVSPLRLKDGSVILVNKGHLPTKLAK 69
+T V+L GV E+LVGPR G PGY VV+P L DG ILVN+G + A+
Sbjct 96 WTRVLLRGVFCHDQEMLVGPRT--KEGQPGYHVVTPFILDDGRRILVNRGWIARSFAE 151
> YGR112w
Length=389
Score = 35.8 bits (81), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 3/56 (5%)
Query 9 EWAYTPVVLHGVLDSSTELLVGPRPGLDVGNPGYCVVSP-LRLKDGSVILVNKGHL 63
+W Y V+L G + E+ VGPR G GY + +P +R G +L+ +G +
Sbjct 127 DWEYRKVILTGHFLHNEEMFVGPRK--KNGEKGYFLFTPFIRDDTGEKVLIERGWI 180
> Hs4507319
Length=300
Score = 35.4 bits (80), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 44/100 (44%), Gaps = 19/100 (19%)
Query 12 YTPVVLHGVLDSSTELLVGPRPGLD-------------VGNPGYCVVSPLRLKD-GSVIL 57
Y PV + G D S EL + PR +D G VV+P D G IL
Sbjct 117 YRPVKVRGCFDHSKELYMMPRTMVDPVREAREGGLISSSTQSGAYVVTPFHCTDLGVTIL 176
Query 58 VNKGHLPTKLAKLPPYPK---SVDENVALLAMVQRQRRQQ 94
VN+G +P K K+ P + ++ V L+ MV+ +Q
Sbjct 177 VNRGFVPRK--KVNPETRQKGQIEGEVDLIGMVRLTETRQ 214
> CE23794
Length=323
Score = 32.0 bits (71), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 31/72 (43%), Gaps = 18/72 (25%)
Query 12 YTPVVLHGVLDSSTELLVGPRPGLDVGNP-----------------GYCVVSPLRLKD-G 53
Y V + G E ++ PR D G G +++P RLK+ G
Sbjct 141 YCRVTVTGEFLHEKEFIISPRGRFDPGKKTSAAAGSMLSENEMSSHGGHLITPFRLKNSG 200
Query 54 SVILVNKGHLPT 65
+IL+N+G LP+
Sbjct 201 KIILINRGWLPS 212
> 7295311
Length=293
Score = 29.6 bits (65), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 33/71 (46%), Gaps = 17/71 (23%)
Query 12 YTPVVLHGVLDSSTELLVGPRP-----------GL----DVGNPGYCVVSPLRLKD-GSV 55
Y V + G E+ +GPR GL D GN GY +V+P +L D +
Sbjct 111 YRLVKIRGRFLHDKEMRLGPRSLIRPDGVETQGGLFSQRDSGN-GYLIVTPFQLADRDDI 169
Query 56 ILVNKGHLPTK 66
+LVN+G + K
Sbjct 170 VLVNRGWVSRK 180
> 7295590
Length=773
Score = 29.3 bits (64), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 1/63 (1%)
Query 73 YPKSVDENVALLAMVQRQRRQQQQRTPLEDVSVERIDHQVRAMQQQRPKQKYVNCGAQQY 132
Y ++ E +A+ A+V RQR +++ + +S E+ A P QK N Q
Sbjct 618 YETTMSEWLAVEAIV-RQREKEKTALAVAKLSAEQARLAANATSSSNPHQKLANGSQQLQ 676
Query 133 LEH 135
LEH
Sbjct 677 LEH 679
> CE23019
Length=648
Score = 28.1 bits (61), Expect = 5.7, Method: Composition-based stats.
Identities = 19/82 (23%), Positives = 45/82 (54%), Gaps = 3/82 (3%)
Query 45 VSPLRLKDGSVILVNKGHL--PTKLAK-LPPYPKSVDENVALLAMVQRQRRQQQQRTPLE 101
++ LR+++ + + L P +++K + KS DE A +R++++ +QR+ LE
Sbjct 511 MAALRVEEVGITASTEAQLMAPEEISKKMKAIEKSKDERDATDKARERRQKKAKQRSMLE 570
Query 102 DVSVERIDHQVRAMQQQRPKQK 123
ER+ + +A++ + ++K
Sbjct 571 KFGEERVFGEQKAVKLAKKERK 592
> 7290389
Length=968
Score = 27.7 bits (60), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 17/39 (43%), Gaps = 4/39 (10%)
Query 23 SSTELLVGPRPGLDVGNPGYCVVSPLRLKDGSVILVNKG 61
SS L P G VGN G C SP G IL N G
Sbjct 921 SSNTLAATPLAGGGVGNSGQCAQSP----SGQFILSNNG 955
Lambda K H
0.316 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1603110344
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40