bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2286_orf1
Length=243
Score E
Sequences producing significant alignments: (Bits) Value
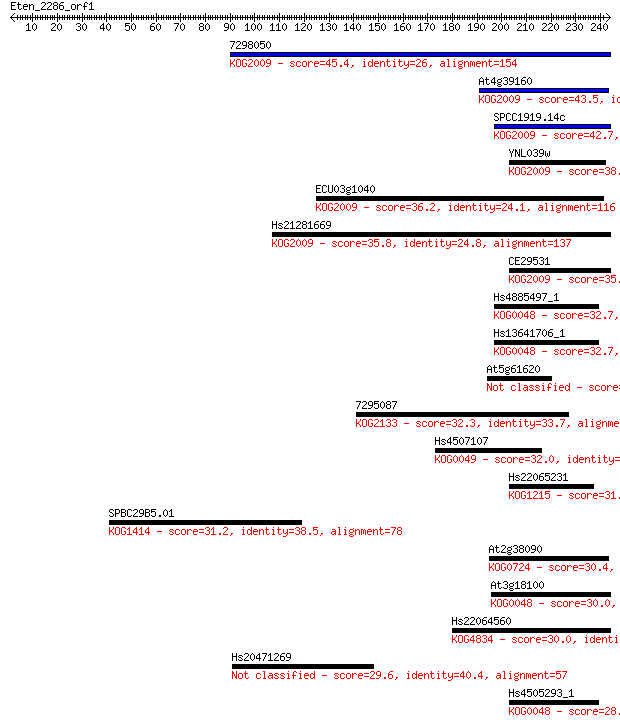
7298050 45.4 1e-04
At4g39160 43.5 4e-04
SPCC1919.14c 42.7 6e-04
YNL039w 38.1 0.016
ECU03g1040 36.2 0.060
Hs21281669 35.8 0.073
CE29531 35.8 0.074
Hs4885497_1 32.7 0.66
Hs13641706_1 32.7 0.66
At5g61620 32.7 0.76
7295087 32.3 0.98
Hs4507107 32.0 1.2
Hs22065231 31.6 1.7
SPBC29B5.01 31.2 2.2
At2g38090 30.4 3.0
At3g18100 30.0 4.3
Hs22064560 30.0 4.7
Hs20471269 29.6 6.3
Hs4505293_1 28.9 9.3
> 7298050
Length=693
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 40/161 (24%), Positives = 70/161 (43%), Gaps = 24/161 (14%)
Query 90 SPQQHQLKQQKQVAGLVSTTNGDGSD--LQGLFEAPAGDSG----VDSLRLDSDGNLVVA 143
+P+ + + ++ V + + GD S + L E P G+S V L+LD++G +++
Sbjct 246 NPENNPMVPKQSVTTIKDESGGDDSKPAVSQLLE-PKGESTSAMLVPQLKLDANGEMIID 304
Query 144 DGPKEVFCCRCLGMSGLLGGGCMCTSGQQRRVLEEDLTSRGAFVLQPYEGAYKKTK-GKR 202
+ E+ + +L + +L ++ T F YK+ K
Sbjct 305 EKTLEIETTAEVEARKVLANSSL--------ILMDETTGDNGF--------YKRHKRTPY 348
Query 203 WTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQLKRKLKR 243
WT +ET +FY L GTD L+ M P + LK K K+
Sbjct 349 WTSDETVRFYRSLQIIGTDFSLMCQMFPTRSRRDLKLKYKK 389
> At4g39160
Length=545
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 30/52 (57%), Gaps = 0/52 (0%)
Query 191 YEGAYKKTKGKRWTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQLKRKLK 242
Y+ KT RW++E+T FYE + G++L +I+ + P T Q+K K K
Sbjct 381 YQTYMNKTSRTRWSKEDTELFYEGIQEFGSNLSMIQQLFPERTREQMKLKFK 432
> SPCC1919.14c
Length=507
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 197 KTKGKRWTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQLKRKLKR 243
+ K ++W +T KFY+ LS GTD LI M P Q+K K K+
Sbjct 362 RQKPEKWNAMDTEKFYKALSQWGTDFALIANMFPTRNRRQIKLKFKQ 408
> YNL039w
Length=594
Score = 38.1 bits (87), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 203 WTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQLKRKL 241
WT EE KFY+ LS GTD LI + P + Q+K K
Sbjct 420 WTVEEMIKFYKALSMWGTDFNLISQLYPYRSRKQVKAKF 458
> ECU03g1040
Length=176
Score = 36.2 bits (82), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 28/119 (23%), Positives = 48/119 (40%), Gaps = 29/119 (24%)
Query 125 GDSGVDSLRLDSDGNLVVADGPKEVFCCRCLGMSGLLGGGCMCTSGQQRRVLEED--LTS 182
D + + + +G++VV D V + + M V+E+D +TS
Sbjct 48 ADQNQEEILMIKNGHIVVNDANMYVDTHKVMEME----------------VMEDDRIVTS 91
Query 183 RGAFVLQPYEGAYKKTKGK-RWTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQLKRK 240
+ K KG RW+++E FY+ L CG + LI ++ P +K K
Sbjct 92 ----------STFSKKKGALRWSKKEIELFYKALEICGIEFSLISSLFPNKERKHIKAK 140
> Hs21281669
Length=2254
Score = 35.8 bits (81), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 34/139 (24%), Positives = 59/139 (42%), Gaps = 22/139 (15%)
Query 107 STTNGDGSDLQGLFEAPAGDSGVDSLRLDSDGNLVVADGPKEVFCCRCLGMSGLLGGGCM 166
ST N + ++++ E G V +++ DG++++ + V R G C
Sbjct 225 STPNAEDNEMEE--ETDDGPLLVPRVKVAEDGSIILDEESLTVEVLRT-------KGPC- 274
Query 167 CTSGQQRRVLEED--LTSRGAFVLQPYEGAYKKTKGKRWTEEETRKFYECLSSCGTDLLL 224
V+EE+ + RG+ Y K K W+ +ET F+ +S GTD +
Sbjct 275 --------VVEENDPIFERGSTTT--YSSFRKNYYSKPWSNKETDMFFLAISMVGTDFSM 324
Query 225 IRTMMPGVTDGQLKRKLKR 243
I + P ++K K KR
Sbjct 325 IGQLFPHRARIEIKNKFKR 343
> CE29531
Length=779
Score = 35.8 bits (81), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 203 WTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQLKRKLKR 243
W+E+ET FYE L G D L+ +P T +LK K R
Sbjct 263 WSEKETDLFYEVLQCTGQDFGLMSHYLPKRTRPELKAKYNR 303
> Hs4885497_1
Length=394
Score = 32.7 bits (73), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 197 KTKGKRWTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQLK 238
+ K WTEEE R Y+ G I ++PG TD +K
Sbjct 141 EVKKTSWTEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIK 182
> Hs13641706_1
Length=394
Score = 32.7 bits (73), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 197 KTKGKRWTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQLK 238
+ K WTEEE R Y+ G I ++PG TD +K
Sbjct 141 EVKKTSWTEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIK 182
> At5g61620
Length=317
Score = 32.7 bits (73), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 14/26 (53%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 194 AYKKTKGKRWTEEETRKFYECLSSCG 219
A++K KGK WTEEE R F L+ G
Sbjct 101 AHEKKKGKPWTEEEHRNFLIGLNKLG 126
> 7295087
Length=1963
Score = 32.3 bits (72), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 40/103 (38%), Gaps = 22/103 (21%)
Query 141 VVADGPKEVFCCRCLGMSGLLGGGCMCTSGQQRRVL---EEDLTSRGAFVLQPYEGAYKK 197
VVADG +F M+ G MC G + L +D T VL ++ Y
Sbjct 32 VVADGDLLMFLRAARSMAAFQG---MCDGGLEDGCLAASRDDTTINALDVL--HDSGYDP 86
Query 198 TKG--------------KRWTEEETRKFYECLSSCGTDLLLIR 226
K K+WTE+ET+KF + L G + I
Sbjct 87 GKALQALVKCPVSKGIDKKWTEDETKKFIKGLRQFGKNFFRIH 129
> Hs4507107
Length=1469
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Query 173 RRVLEEDLTSRGAF-VLQPYEGAYKKTKGKRWTEEETRKFYECL 215
+++ EE TSR AF LQ ++ K K K WTEEE R + +
Sbjct 318 QKIAEELGTSRSAFQCLQKFQQHNKALKRKEWTEEEDRMLTQLV 361
> Hs22065231
Length=1535
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 11/34 (32%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 203 WTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQ 236
WT+ +TR + G++++L+R+ +PG+ D Q
Sbjct 897 WTDWQTRSIHRADKGTGSNVILVRSNLPGLMDMQ 930
> SPBC29B5.01
Length=566
Score = 31.2 bits (69), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 30/104 (28%), Positives = 40/104 (38%), Gaps = 34/104 (32%)
Query 41 LWDLVKR--NLRRKTAAGAAARRSAGTGIATGTVESPVDHTEDPFALLLTPS-------- 90
++D R L + A AAAR S+GTG G E PF LLTP+
Sbjct 175 IYDATLRPDYLNNPSDASAAARFSSGTGFTPGVNE--------PFRSLLTPTGAGFPAPS 226
Query 91 ----------------PQQHQLKQQKQVAGLVSTTNGDGSDLQG 118
P Q++ + +V+ TNGD SD G
Sbjct 227 PGTANLLGFHTFDSQFPDQYRFTPRDGKPPVVNGTNGDQSDYFG 270
> At2g38090
Length=291
Score = 30.4 bits (67), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query 195 YKKTKGKRWTEEETRKFYECLSSCGTD----LLLIRTMMPGVTDGQLKRKLK 242
+++ +G +WT EE +KF L+ D + M+PG T G + ++ +
Sbjct 14 FQENRGTKWTAEENKKFENALAFYDKDTPDRWSRVAAMLPGKTVGDVIKQYR 65
> At3g18100
Length=791
Score = 30.0 bits (66), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 24/48 (50%), Gaps = 1/48 (2%)
Query 196 KKTKGKRWTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQLKRKLKR 243
K +GK WTEEE K E ++ G + T + TD Q R+ KR
Sbjct 539 KVNRGK-WTEEEDEKLREAIAEHGYSWSKVATNLSCRTDNQCLRRWKR 585
> Hs22064560
Length=172
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 26/64 (40%), Gaps = 0/64 (0%)
Query 180 LTSRGAFVLQPYEGAYKKTKGKRWTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQLKR 239
T G +Q + A G +WTE E + G DL I ++ T Q+K
Sbjct 19 FTKLGELTMQLHPVADSSPAGAKWTETEIEMLRAAVKRFGDDLNHISCVIKERTVAQIKA 78
Query 240 KLKR 243
+KR
Sbjct 79 TVKR 82
> Hs20471269
Length=431
Score = 29.6 bits (65), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 30/60 (50%), Gaps = 3/60 (5%)
Query 91 PQQHQLKQQKQVAGLVSTTNGDGSDLQGL-FEAPAGDSGVDSLRLDSDG--NLVVADGPK 147
PQ+ QL + L + S L+G + AGDSG+DS R S G N V+ DG K
Sbjct 340 PQKFQLFNTSHMPVLAQDVQYEHSHLEGTENHSMAGDSGIDSPRTQSLGSNNSVILDGLK 399
> Hs4505293_1
Length=313
Score = 28.9 bits (63), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 17/36 (47%), Gaps = 0/36 (0%)
Query 203 WTEEETRKFYECLSSCGTDLLLIRTMMPGVTDGQLK 238
WTEEE R E G I M+PG TD +K
Sbjct 138 WTEEEDRIICEAHKVLGNRWAEIAKMLPGRTDNAVK 173
Lambda K H
0.315 0.131 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4867742342
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40