bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2186_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
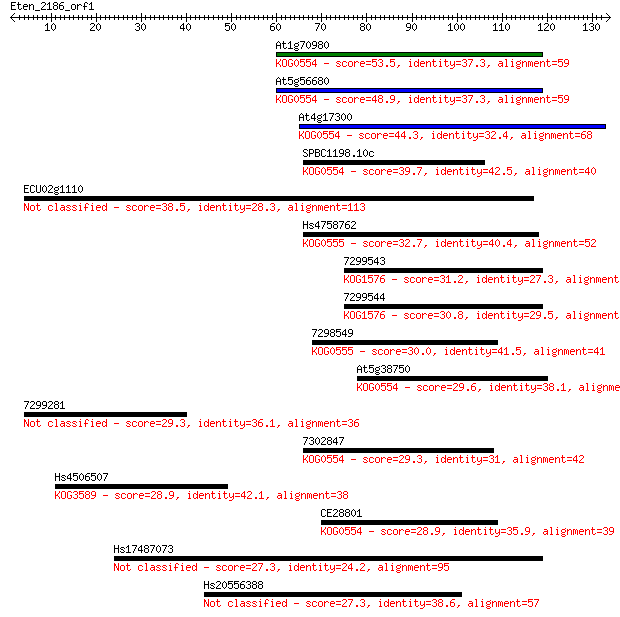
At1g70980 53.5 1e-07
At5g56680 48.9 2e-06
At4g17300 44.3 7e-05
SPBC1198.10c 39.7 0.002
ECU02g1110 38.5 0.004
Hs4758762 32.7 0.18
7299543 31.2 0.49
7299544 30.8 0.66
7298549 30.0 1.2
At5g38750 29.6 1.5
7299281 29.3 2.0
7302847 29.3 2.0
Hs4506507 28.9 2.9
CE28801 28.9 3.1
Hs17487073 27.3 7.7
Hs20556388 27.3 7.7
> At1g70980
Length=571
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 60 AAATFIGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCG 118
A G++V + GW ++ R+QG G+ F+ + DGSC ANLQV+V+ ++ L+ G
Sbjct 41 GGAKLAGQKVRIGGWVKTGRQQGKGTFAFLEVNDGSCPANLQVMVDSSLYDLSRLVATG 99
> At5g56680
Length=572
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 60 AAATFIGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCG 118
A G+ V + GW +S R QG + F+ + DGSC ANLQV+V+ ++ L+ G
Sbjct 44 GGAGLAGQTVRIGGWVKSGRDQGKRTFSFLAVNDGSCPANLQVMVDPSLYDVSNLVATG 102
> At4g17300
Length=567
Score = 44.3 bits (103), Expect = 7e-05, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 38/70 (54%), Gaps = 4/70 (5%)
Query 65 IGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCGV--GCS 122
+G+ + + GW R++R Q S+ F+ + DGSC +NLQ V+ +D++ + G S
Sbjct 108 VGQSLNIMGWVRTLRSQS--SVTFIEINDGSCLSNLQCVMTSDAEGYDQVESGSILTGAS 165
Query 123 FKFCGRVVES 132
G +V S
Sbjct 166 VSVQGTIVAS 175
> SPBC1198.10c
Length=441
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 17/40 (42%), Positives = 27/40 (67%), Gaps = 2/40 (5%)
Query 66 GRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVN 105
G +++ GW RSIRK ++CF +++DG+C LQVV +
Sbjct 20 GELISINGWVRSIRKLK--NVCFAMVSDGTCQQALQVVTS 57
> ECU02g1110
Length=835
Score = 38.5 bits (88), Expect = 0.004, Method: Composition-based stats.
Identities = 32/115 (27%), Positives = 55/115 (47%), Gaps = 6/115 (5%)
Query 4 CCFLSHLFT-MTMAGSAPSPPALPPVSETAALSYVQPIAVYGPGGGRM-RLAALLASCAA 61
CC L + +T M M APS + E + + P+ P GG++ + +LLA
Sbjct 200 CCDLLYNYTSMEMKAEAPSEKTFEYIMEKLLVFFEIPL----PRGGKLLKTFSLLAEKDL 255
Query 62 ATFIGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIK 116
FIG + + S S++K+ LC + L + ++ V + IP+F +LI+
Sbjct 256 GAFIGSGMRLMQLSASLKKEIVRELCELNLKNEQIYEFIRSVRHILIPEFIDLIR 310
> Hs4758762
Length=548
Score = 32.7 bits (73), Expect = 0.18, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 30/52 (57%), Gaps = 9/52 (17%)
Query 66 GRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKC 117
G++V V GW +R+Q G +L F+VL DG+ LQ V+ DEL +C
Sbjct 125 GQRVKVFGWVHRLRRQ-GKNLMFLVLRDGT--GYLQCVLA------DELCQC 167
> 7299543
Length=345
Score = 31.2 bits (69), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 12/44 (27%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 75 SRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCG 118
+S++ G + + + D A +L +V+N+T+P ++L+K G
Sbjct 126 EKSLKLLGLDYVDVIQIHDIEFAKDLDIVINETLPTLEQLVKEG 169
> 7299544
Length=294
Score = 30.8 bits (68), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 75 SRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCG 118
RS+ + + + + D A NL +V+N+TIP +E ++ G
Sbjct 99 KRSLERLQLDRVDILQVHDVDAAPNLDIVLNETIPVLEEYVQAG 142
> 7298549
Length=558
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 24/41 (58%), Gaps = 3/41 (7%)
Query 68 QVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTI 108
+V V GW +R+Q G SL F+ L DG+ LQ V+N +
Sbjct 137 RVKVYGWVHRLRRQ-GKSLIFITLRDGT--GFLQCVLNDQL 174
> At5g38750
Length=135
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 78 IRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCGV 119
I ++ G S F+ + GSCAANLQV + E+I+ V
Sbjct 31 ISRELGKSFTFLEVNYGSCAANLQVKIPLQGKATKEMIELNV 72
> 7299281
Length=1336
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query 4 CCFLSHLFTMTMAGSAPSPPALPPVSETAALSYVQP 39
C +H + + A AP PPA P+ ++A +YV P
Sbjct 1012 CSSQAH-YHASTAEEAPKPPAERPLQKSATSTYVSP 1046
> 7302847
Length=460
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 25/42 (59%), Gaps = 2/42 (4%)
Query 66 GRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKT 107
G + + GW +++R+ + F+ + DGS ++ QVVV +T
Sbjct 24 GDSLAIQGWIKNVRRLKNNT--FLDINDGSTSSRFQVVVPRT 63
> Hs4506507
Length=446
Score = 28.9 bits (63), Expect = 2.9, Method: Composition-based stats.
Identities = 16/38 (42%), Positives = 19/38 (50%), Gaps = 8/38 (21%)
Query 11 FTMTMAGSAPSPPALPPVSETAALSYVQPIAVYGPGGG 48
FT S+PSP LP + V+P A GPGGG
Sbjct 413 FTWRPRHSSPSPALLP--------TPVEPTAACGPGGG 442
> CE28801
Length=448
Score = 28.9 bits (63), Expect = 3.1, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Query 70 TVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTI 108
T+ GW++S++K G FV + G +Q+V NK I
Sbjct 14 TLDGWAKSVQKSGKN--VFVKIDKGRAGEPIQLVANKEI 50
> Hs17487073
Length=120
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 23/99 (23%), Positives = 38/99 (38%), Gaps = 5/99 (5%)
Query 24 ALPPVSETAALSYVQPIAVYGPGG----GRMRLAALLASCAAATFIGRQVTVCGWSRSIR 79
A PP+ E A L Q + GP G + LA + + +G +R
Sbjct 22 AHPPLEERARLLRGQSVQQVGPQGLLYVQQRELAVTSPKDGSISILGSDDATTCHIVVLR 81
Query 80 KQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCG 118
G G+ C A + +++N +I F + +CG
Sbjct 82 HTGNGATCLTHCDGTDTKAEVPLIMN-SIKSFSDHAQCG 119
> Hs20556388
Length=424
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 28/57 (49%), Gaps = 2/57 (3%)
Query 44 GPGGGRMRLAALLASCAAATFIGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANL 100
G G R L SCA F GRQ+ + G R R G G+ C+V +T G A +L
Sbjct 278 GKSGRRDHQLYLPVSCALFAFSGRQLRL-GEFREFRA-GPGTPCYVDVTYGYPAGDL 332
Lambda K H
0.325 0.138 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1356426142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40