bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2159_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
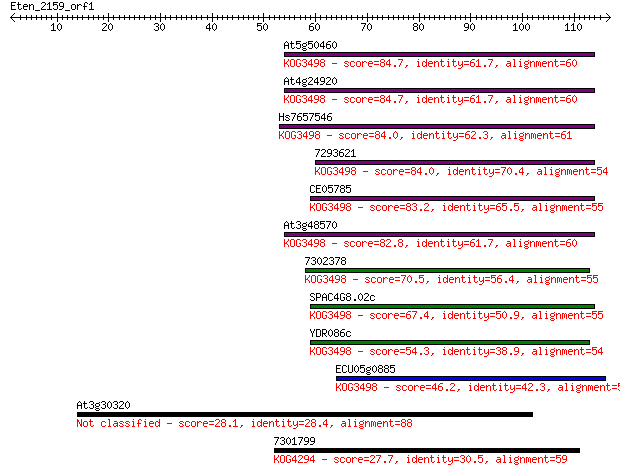
At5g50460 84.7 4e-17
At4g24920 84.7 4e-17
Hs7657546 84.0 6e-17
7293621 84.0 7e-17
CE05785 83.2 1e-16
At3g48570 82.8 2e-16
7302378 70.5 7e-13
SPAC4G8.02c 67.4 6e-12
YDR086c 54.3 5e-08
ECU05g0885 46.2 1e-05
At3g30320 28.1 4.2
7301799 27.7 5.0
> At5g50460
Length=69
Score = 84.7 bits (208), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 37/60 (61%), Positives = 48/60 (80%), Gaps = 0/60 (0%)
Query 54 VTELADFGADSVRLVRRCTKPDAREFKKIAWACAVGFLLMGFIGYVVKLVFIPINNILVG 113
V L DF DS+RLV+RC KPD +EF K+A A+GF++MGF+G+ VKL+FIPINNI+VG
Sbjct 8 VDPLRDFAKDSIRLVKRCHKPDRKEFTKVAVRTAIGFVVMGFVGFFVKLIFIPINNIIVG 67
> At4g24920
Length=69
Score = 84.7 bits (208), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 37/60 (61%), Positives = 48/60 (80%), Gaps = 0/60 (0%)
Query 54 VTELADFGADSVRLVRRCTKPDAREFKKIAWACAVGFLLMGFIGYVVKLVFIPINNILVG 113
V L DF DS+RLV+RC KPD +EF K+A A+GF++MGF+G+ VKL+FIPINNI+VG
Sbjct 8 VDPLRDFAKDSIRLVKRCHKPDRKEFTKVAVRTAIGFVVMGFVGFFVKLIFIPINNIIVG 67
> Hs7657546
Length=68
Score = 84.0 bits (206), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 38/61 (62%), Positives = 48/61 (78%), Gaps = 0/61 (0%)
Query 53 WVTELADFGADSVRLVRRCTKPDAREFKKIAWACAVGFLLMGFIGYVVKLVFIPINNILV 112
+V F DS+RLV+RCTKPD +EF+KIA A A+GF +MGFIG+ VKL+ IPINNI+V
Sbjct 7 FVEPSRQFVKDSIRLVKRCTKPDRKEFQKIAMATAIGFAIMGFIGFFVKLIHIPINNIIV 66
Query 113 G 113
G
Sbjct 67 G 67
> 7293621
Length=68
Score = 84.0 bits (206), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 38/54 (70%), Positives = 46/54 (85%), Gaps = 0/54 (0%)
Query 60 FGADSVRLVRRCTKPDAREFKKIAWACAVGFLLMGFIGYVVKLVFIPINNILVG 113
F DS+RLV+RCTKPD +EF+KIA A AVGF +MGFIG+ VKL+ IPINNI+VG
Sbjct 14 FAKDSIRLVKRCTKPDRKEFQKIAIATAVGFCIMGFIGFFVKLIHIPINNIIVG 67
> CE05785
Length=68
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 36/55 (65%), Positives = 45/55 (81%), Gaps = 0/55 (0%)
Query 59 DFGADSVRLVRRCTKPDAREFKKIAWACAVGFLLMGFIGYVVKLVFIPINNILVG 113
F DS RLV+RCTKPD +E++KIA A A+GF +MGFIG+ VKL+ IPINNI+VG
Sbjct 13 QFSKDSYRLVKRCTKPDRKEYQKIAMATAIGFAIMGFIGFFVKLIHIPINNIIVG 67
> At3g48570
Length=69
Score = 82.8 bits (203), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 37/60 (61%), Positives = 47/60 (78%), Gaps = 0/60 (0%)
Query 54 VTELADFGADSVRLVRRCTKPDAREFKKIAWACAVGFLLMGFIGYVVKLVFIPINNILVG 113
+ L DF SVRLV+RC KPD +EF K+A A+GF++MGF+G+ VKLVFIPINNI+VG
Sbjct 8 IDPLRDFAKSSVRLVQRCHKPDRKEFTKVAVRTAIGFVVMGFVGFFVKLVFIPINNIIVG 67
> 7302378
Length=105
Score = 70.5 bits (171), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 42/55 (76%), Gaps = 0/55 (0%)
Query 58 ADFGADSVRLVRRCTKPDAREFKKIAWACAVGFLLMGFIGYVVKLVFIPINNILV 112
DF +S+R +RCTKPD REF++I+ VGFL+MG IG+VVKL+ IPI NI++
Sbjct 50 KDFYKNSLRFYKRCTKPDRREFQRISIGIGVGFLIMGLIGFVVKLMHIPIVNIIM 104
> SPAC4G8.02c
Length=70
Score = 67.4 bits (163), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 28/55 (50%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 59 DFGADSVRLVRRCTKPDAREFKKIAWACAVGFLLMGFIGYVVKLVFIPINNILVG 113
+F + ++RC KPD +EF I+ A A GF+LMG IGY++KL+ IPIN +LVG
Sbjct 14 NFYKEGSHFIKRCVKPDRKEFLSISKAVATGFVLMGLIGYIIKLIHIPINKVLVG 68
> YDR086c
Length=80
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 35/54 (64%), Gaps = 0/54 (0%)
Query 59 DFGADSVRLVRRCTKPDAREFKKIAWACAVGFLLMGFIGYVVKLVFIPINNILV 112
+F + + + +C KPD +E+ KI A +GF+ +G IGY +KL+ IPI ++V
Sbjct 27 EFVREGTQFLAKCKKPDLKEYTKIVKAVGIGFIAVGIIGYAIKLIHIPIRYVIV 80
> ECU05g0885
Length=72
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 33/54 (61%), Gaps = 2/54 (3%)
Query 64 SVRL-VRRCTKPDAREFKKIAWACAVGFLLMGFIGYVVKLVFIPINNILV-GPG 115
S+RL ++C +P +E A+G +G +GY +KL+ IPINNI+V PG
Sbjct 17 SIRLFSKKCVRPSGKELSMSIKRHAIGIGFLGILGYAIKLIHIPINNIIVSSPG 70
> At3g30320
Length=695
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 25/88 (28%), Positives = 39/88 (44%), Gaps = 10/88 (11%)
Query 14 VPLRDASGCCAAAADTLHKMVQLNKVPSFLLDRSHPLGFWVTELADFGADSVRLVRRCTK 73
V LR+ CC D L + + + + P+G W+TEL +RL++R
Sbjct 412 VVLRNPINCCE---DYLFEKYVTGVLQTVYQGKRRPIGSWITELG-----VMRLLQRV-- 461
Query 74 PDAREFKKIAWACAVGFLLMGFIGYVVK 101
P R + I+ GF + G + Y VK
Sbjct 462 PLMRFQEIISMFKEFGFPVQGQLRYFVK 489
> 7301799
Length=548
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 31/60 (51%), Gaps = 6/60 (10%)
Query 52 FWVTELADFGADS-VRLVRRCTKPDAREFKKIAWACAVGFLLMGFIGYVVKLVFIPINNI 110
F + L D+ A+S V +R P R+F+++ W LL IG +V V++ +N +
Sbjct 43 FMLPYLKDYAAESSVHGIRYLADPKMRKFERVIWLL---ILLTTSIGAIV--VYVDLNEL 97
Lambda K H
0.329 0.142 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174970866
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40