bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2128_orf1
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
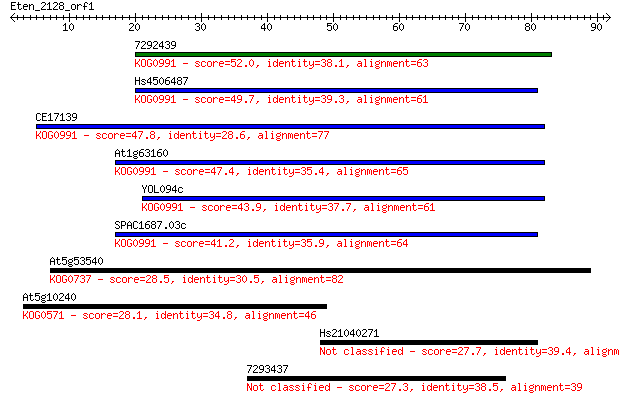
7292439 52.0 3e-07
Hs4506487 49.7 1e-06
CE17139 47.8 4e-06
At1g63160 47.4 7e-06
YOL094c 43.9 7e-05
SPAC1687.03c 41.2 5e-04
At5g53540 28.5 3.2
At5g10240 28.1 4.4
Hs21040271 27.7 5.9
7293437 27.3 7.1
> 7292439
Length=331
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 24/63 (38%), Positives = 41/63 (65%), Gaps = 2/63 (3%)
Query 20 LGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLAQLC 79
LGY+P D++ V +R + + EHL L+F+ +G+THM + G+++ LQ+ +LA+LC
Sbjct 268 LGYSPEDIIANIFRVCKRIN--IDEHLKLDFIREIGITHMKIIDGINSLLQLTALLAKLC 325
Query 80 KVA 82
A
Sbjct 326 IAA 328
> Hs4506487
Length=354
Score = 49.7 bits (117), Expect = 1e-06, Method: Composition-based stats.
Identities = 24/61 (39%), Positives = 40/61 (65%), Gaps = 2/61 (3%)
Query 20 LGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLAQLC 79
LGY+P D++ V + ++ E+L LEF+ +G THM +A G+++ LQM +LA+LC
Sbjct 288 LGYSPEDIIGNIFRVCKTF--QMAEYLKLEFIKEIGYTHMKIAEGVNSLLQMAGLLARLC 345
Query 80 K 80
+
Sbjct 346 Q 346
> CE17139
Length=334
Score = 47.8 bits (112), Expect = 4e-06, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 46/79 (58%), Gaps = 2/79 (2%)
Query 5 RVWYGAQSVARVILSLGYTPLDVVTTFRGVLRRAD--SELQEHLLLEFLGIVGMTHMTMA 62
R ++ A + LG++ D+V+T V++ + + E L +E++ + M HM +
Sbjct 247 RKFFEASKIIHEFHRLGFSSDDIVSTLFRVVKTVELSKNVSEQLRMEYIRQIAMCHMRIV 306
Query 63 GGLSTELQMEKMLAQLCKV 81
GL+++LQ+ +++A LC+V
Sbjct 307 QGLTSKLQLSRLIADLCRV 325
> At1g63160
Length=333
Score = 47.4 bits (111), Expect = 7e-06, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 39/65 (60%), Gaps = 2/65 (3%)
Query 17 ILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLA 76
+ LGY+P D++TT +++ D + E+L LEF+ G HM + G+ + LQ+ +LA
Sbjct 264 LYDLGYSPTDIITTLFRIIKNYD--MAEYLKLEFMKETGFAHMRICDGVGSYLQLCGLLA 321
Query 77 QLCKV 81
+L V
Sbjct 322 KLSIV 326
> YOL094c
Length=323
Score = 43.9 bits (102), Expect = 7e-05, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 42/62 (67%), Gaps = 3/62 (4%)
Query 21 GYTPLDVVTT-FRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLAQLC 79
GY+ +D+VTT FR + + ++++E + LE + +G+THM + G+ T LQ+ MLA++
Sbjct 260 GYSSIDIVTTSFR--VTKNLAQVKESVRLEMIKEIGLTHMRILEGVGTYLQLASMLAKIH 317
Query 80 KV 81
K+
Sbjct 318 KL 319
> SPAC1687.03c
Length=342
Score = 41.2 bits (95), Expect = 5e-04, Method: Composition-based stats.
Identities = 23/64 (35%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 17 ILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLA 76
I LG++ +D+VT V++ DS + E LE L +G THM + G+ T LQ+ ++
Sbjct 270 IWDLGFSAVDIVTNMFRVVKTMDS-IPEFSRLEMLKEIGQTHMIILEGVQTLLQLSGLVC 328
Query 77 QLCK 80
+L K
Sbjct 329 RLAK 332
> At5g53540
Length=403
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 39/89 (43%), Gaps = 20/89 (22%)
Query 7 WYG-AQSVARVILSLGYT--P----LDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHM 59
W+G AQ + + SL Y P +D V +F G R D+E ++ EF M
Sbjct 162 WFGDAQKLVSAVFSLAYKLQPAIIFIDEVDSFLGQRRSTDNEAMSNMKTEF--------M 213
Query 60 TMAGGLSTELQMEKMLAQLCKVAISFRPS 88
+ G +T+ M+ +A + RPS
Sbjct 214 ALWDGFTTDQNARVMV-----LAATNRPS 237
> At5g10240
Length=578
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Query 3 GFRVWYGAQSVARVILSLGYTPLDVVTTF-RGVLRRADSELQEHLLL 48
G R WY + V+ S Y PL V TF + V++R +++ +LL
Sbjct 186 GLRRWYNPPWFSEVVPSTPYDPLVVRNTFEKAVIKRLMTDVPFGVLL 232
> Hs21040271
Length=980
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 48 LEFLGIVGMTHMTMAGGLSTELQMEKMLAQLCK 80
L F+G +TH+T+AG + E M ML L +
Sbjct 725 LAFIGKKTLTHLTLAGHIEWERTMMLMLCDLLR 757
> 7293437
Length=161
Score = 27.3 bits (59), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 37 RADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKML 75
R S Q L F+GI G M M G+ ELQ+E ++
Sbjct 113 RTSSPTQTLDFLIFIGIPGDVEMAMEMGMDMELQIEMVM 151
Lambda K H
0.326 0.139 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174483934
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40