bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
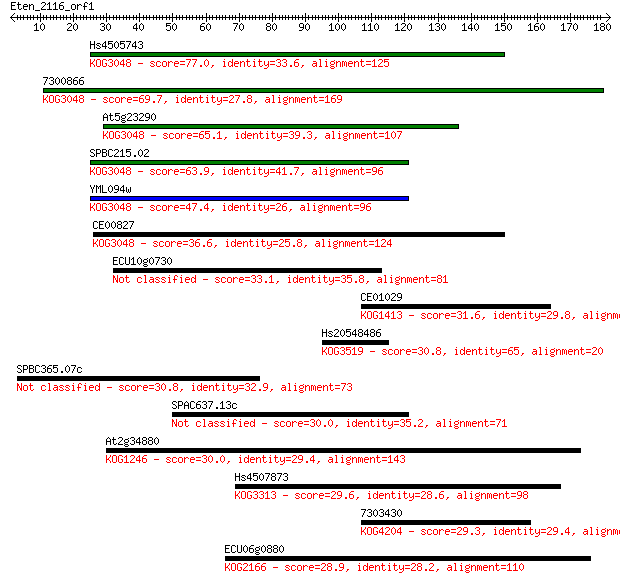
Query= Eten_2116_orf1
Length=181
Score E
Sequences producing significant alignments: (Bits) Value
Hs4505743 77.0 2e-14
7300866 69.7 3e-12
At5g23290 65.1 7e-11
SPBC215.02 63.9 2e-10
YML094w 47.4 2e-05
CE00827 36.6 0.030
ECU10g0730 33.1 0.29
CE01029 31.6 0.83
Hs20548486 30.8 1.4
SPBC365.07c 30.8 1.5
SPAC637.13c 30.0 2.3
At2g34880 30.0 2.5
Hs4507873 29.6 3.1
7303430 29.3 4.5
ECU06g0880 28.9 6.4
> Hs4505743
Length=167
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 42/125 (33%), Positives = 72/125 (57%), Gaps = 6/125 (4%)
Query 25 NLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEA 84
N+ L L Q L + + EVE LS ++QLK ++ EA++ + L+ K E
Sbjct 19 NITELNLPQLEMLKNQLDQEVEFLSTSIAQLKVVQTKYVEAKDCLNVLN------KSNEG 72
Query 85 PEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEK 144
E+LVPLT ++YV G+L + VL+D+G GY ++KT +D K R + F+++Q +++
Sbjct 73 KELLVPLTSSMYVPGKLHDVEHVLIDVGTGYYVEKTAEDAKDFFKRKIDFLTKQMEKIQP 132
Query 145 IISDK 149
+ +K
Sbjct 133 ALQEK 137
> 7300866
Length=168
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 47/169 (27%), Positives = 86/169 (50%), Gaps = 6/169 (3%)
Query 11 MAAPTAGTVKQDINNLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVI 70
MAA + K ++ +L L+ +Q + + E E+ N+ + LS L ++ + ++EA+
Sbjct 1 MAATPSAPAKSEMIDLTKLSPEQLIQIKQEFEQEITNVQDSLSTLHGCQAKYAGSKEALG 60
Query 71 QLDAYRQRQKEGEAPEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMR 130
Q E ++LVPLT ++YV GR+ ++ ++DIG GY I+K + K R
Sbjct 61 TF------QPNWENRQILVPLTSSMYVPGRVKDLNRFVIDIGTGYYIEKDLEGSKDYFKR 114
Query 131 ILTFMSEQQARLEKIISDKVKQLQVLVATLNRSRDEAPSGASVARRQPA 179
+ ++ EQ ++EKI K + +++ L + A S + QPA
Sbjct 115 RVEYVQEQIEKIEKIHLQKTRFYNSVMSVLEMKQAAAAKLQSQQQSQPA 163
> At5g23290
Length=151
Score = 65.1 bits (157), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 42/107 (39%), Positives = 59/107 (55%), Gaps = 6/107 (5%)
Query 29 LTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEAPEVL 88
+ +DQ +L +A+ EV L + L+ ++ A R+ A A+ L Q +K +L
Sbjct 13 MGIDQLKALKEQADLEVNLLQDSLNNIRTATVRLDAAAAALNDLSLRPQGKK------ML 66
Query 89 VPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFM 135
VPLT +LYV G LD DKVLVDIG GY I+KT D K R + +
Sbjct 67 VPLTASLYVPGTLDEADKVLVDIGTGYFIEKTMDDGKDYCQRKINLL 113
> SPBC215.02
Length=154
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 40/97 (41%), Positives = 60/97 (61%), Gaps = 6/97 (6%)
Query 25 NLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEA 84
+L SL+L+Q S ++++ ++E+E LS QL A + E V DA R E +
Sbjct 9 DLTSLSLEQLSEVIKQLDSELEYLSTSYGQLGRAQLKFRECLANVN--DAVR---AENDG 63
Query 85 PEVLVPLTGALYVKGRLDC-DDKVLVDIGAGYMIQKT 120
EVLVPLT +LYV G+L+ + K+LVDIG GY ++K+
Sbjct 64 KEVLVPLTSSLYVPGKLNLGNSKLLVDIGTGYYVEKS 100
> YML094w
Length=163
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 52/96 (54%), Gaps = 5/96 (5%)
Query 25 NLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEA 84
+L L +Q +++ ++ + E+++ + L L A + +E + D Q E
Sbjct 7 DLTKLNPEQLNAVKQQFDQELQHFTQSLQALTMAKGKFTECID-----DIKTVSQAGNEG 61
Query 85 PEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKT 120
++LVP + +LY+ G++ + K +VDIG GY ++K+
Sbjct 62 QKLLVPASASLYIPGKIVDNKKFMVDIGTGYYVEKS 97
> CE00827
Length=152
Score = 36.6 bits (83), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 56/124 (45%), Gaps = 6/124 (4%)
Query 26 LGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEAP 85
L L+L Q L + E E+ + LK +R +++ LD + A
Sbjct 10 LSELSLQQLGELQKNCEQELNFFQESFNALKGLLTR---NEKSISALDDVKIATAGHTA- 65
Query 86 EVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEKI 145
L+PL+ +LY++ L K LV+IG GY ++ + K R +++Q +E I
Sbjct 66 --LIPLSESLYIRAELSDPSKHLVEIGTGYFVELDREKAKAIFDRKKEHITKQVETVEGI 123
Query 146 ISDK 149
+ +K
Sbjct 124 LKEK 127
> ECU10g0730
Length=266
Score = 33.1 bits (74), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 29/88 (32%), Positives = 42/88 (47%), Gaps = 13/88 (14%)
Query 32 DQCSSLV------RRAEAEVE-NLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEA 84
D C SLV R+ +A + NL +L +F E A+ Q+D Y + +G
Sbjct 173 DNCCSLVVEDVNGRKVDARISLNLGRYLGLREFN----REVLSALPQMDFYWKGTTDGR- 227
Query 85 PEVLVPLTGALYVKGRLDCDDKVLVDIG 112
EVLV A+YV G+L D L+ +G
Sbjct 228 -EVLVQKNLAVYVDGKLRRDATFLMKLG 254
> CE01029
Length=437
Score = 31.6 bits (70), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 107 VLVDIGAGYMIQKTYQDEK--KDAMRILTFMSEQQARLEKIISDKVKQLQVLVATLNRS 163
VL+ + IQ + ++ K+ +R+L M E +RLEK++ + + ++ L T+NR+
Sbjct 12 VLIYFSWNFYIQNGFTNKAIVKEELRLLKEMDEAGSRLEKLLKVEKEHMERLSETMNRA 70
> Hs20548486
Length=652
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 13/20 (65%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 95 LYVKGRLDCDDKVLVDIGAG 114
LY KGRLD D+ LVD+G G
Sbjct 496 LYYKGRLDMDEMELVDLGDG 515
> SPBC365.07c
Length=547
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 46/78 (58%), Gaps = 10/78 (12%)
Query 3 RSFMLCEAMA-APTAGT---VKQDINNLGSLTL-DQCSSLVRRAEAEVENLSNHLSQLKF 57
R F L E+ A +PT+GT +++ N+ ++L +Q ++ +RR EAE++ ++QL
Sbjct 409 RKFSLYESEAISPTSGTPSNLEKGAGNVPDVSLLEQLATTIRRLEAELQTTKQQVAQLII 468
Query 58 AASRVSEAREAVIQLDAY 75
+ +AR+ ++ DAY
Sbjct 469 ---QRDQARQEIV--DAY 481
> SPAC637.13c
Length=498
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 37/74 (50%), Gaps = 13/74 (17%)
Query 50 NHLSQLKFAASRVSEAREAVIQLDAYRQRQKE---GEAPEVLVPLTGALYVKGRLDCDDK 106
NHL ++ RVSEA+ + Y +R+ E G P V A+ G+ +D+
Sbjct 248 NHLQEV----VRVSEAQTLAGEWTGYAKREPEFIHGSVPPRSV---DAIKYPGK---NDQ 297
Query 107 VLVDIGAGYMIQKT 120
V I AGY+I+KT
Sbjct 298 PTVPIMAGYLIRKT 311
> At2g34880
Length=806
Score = 30.0 bits (66), Expect = 2.5, Method: Composition-based stats.
Identities = 42/168 (25%), Positives = 67/168 (39%), Gaps = 41/168 (24%)
Query 30 TLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEAPEVLV 89
T+D+ SSLVR E E ++L LS++ S + + I + KE + E
Sbjct 559 TIDELSSLVRALEGESDDLKAWLSKVMEGCSETQKGESSGIIV-------KEKQVQEECF 611
Query 90 PLTGALYVKGRLDCDDKVLVDIGA--------GYMI------------QKTYQDEKK--- 126
L G + C+D ++D+ A G+++ K ++ K
Sbjct 612 DLNGECNKSSEI-CEDASIMDLAAYHVEPINLGFLVVGKLWCNKHAIFPKGFKSRVKFYN 670
Query 127 --DAMRILTFMSEQQARLEKIISDKVKQLQVLVATLNRSRDEAPSGAS 172
D MRI ++SE I D + TL S+DE+ S AS
Sbjct 671 VQDPMRISYYVSE--------IVDAGLLGPLFKVTLEESQDESFSYAS 710
> Hs4507873
Length=197
Score = 29.6 bits (65), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 43/99 (43%), Gaps = 15/99 (15%)
Query 69 VIQLDAYRQRQKEG-EAPEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKD 127
+++ Y Q++KE + E L LY K + DKV + +GA M++
Sbjct 84 TLEILKYMQKKKESTNSMETRFLLADNLYCKASVPPTDKVCLWLGANVMLEYD------- 136
Query 128 AMRILTFMSEQQARLEKIISDKVKQLQVLVATLNRSRDE 166
+ E QA LEK +S K L L L+ RD+
Sbjct 137 -------IDEAQALLEKNLSTATKNLDSLEEDLDFLRDQ 168
> 7303430
Length=2061
Score = 29.3 bits (64), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Query 107 VLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEKIISDKVKQLQVLV 157
+L ++ G M T++D ++ I ++S L+K++S+ V+QLQ V
Sbjct 1566 MLKNVLDGNMDSNTFEDTMREMFGIYAYIS---FTLDKVVSNAVRQLQYCV 1613
> ECU06g0880
Length=690
Score = 28.9 bits (63), Expect = 6.4, Method: Composition-based stats.
Identities = 31/121 (25%), Positives = 55/121 (45%), Gaps = 11/121 (9%)
Query 66 REAVIQ-LDAYRQRQKEGEAPEVLVP-----LTGALYVKGRLDCDDKVLVDIGAGYMIQK 119
RE V++ L+ RQ ++ + LVP + L++ GRL +D+V DI +
Sbjct 299 RELVVEYLNICRQMEENLMREDELVPYLREEIRKHLHMSGRLGFEDEVYRDIDRTVRNGE 358
Query 120 TYQDEKKDAMRILTFMSEQQARLEKIISDKVKQLQVLVATLNRSR-----DEAPSGASVA 174
E + + ++ MS ++ + KI D ++L + L R E GAS++
Sbjct 359 EMGGEMEALVMFVSLMSSKEVIVGKIARDARRRLMSCKSNLKREEMLVLSMEKHLGASIS 418
Query 175 R 175
R
Sbjct 419 R 419
Lambda K H
0.316 0.129 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2842629034
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40