bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
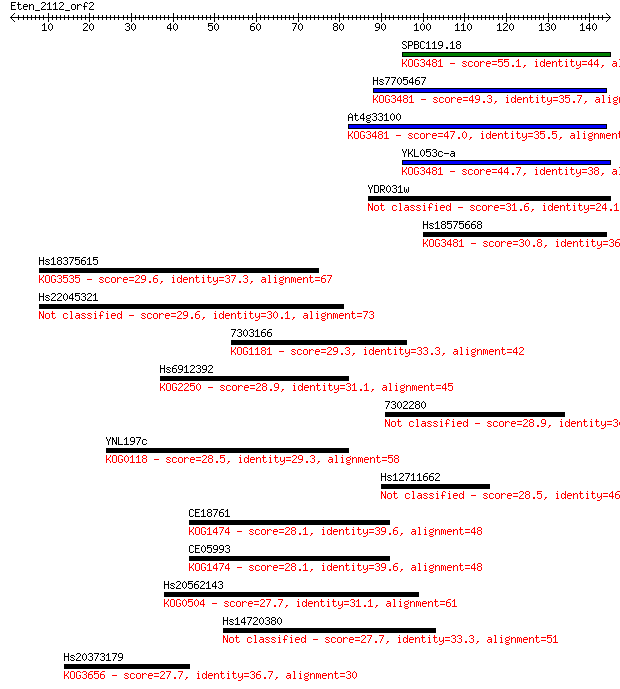
Query= Eten_2112_orf2
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
SPBC119.18 55.1 4e-08
Hs7705467 49.3 2e-06
At4g33100 47.0 1e-05
YKL053c-a 44.7 6e-05
YDR031w 31.6 0.51
Hs18575668 30.8 0.90
Hs18375615 29.6 1.8
Hs22045321 29.6 2.1
7303166 29.3 2.6
Hs6912392 28.9 3.1
7302280 28.9 3.9
YNL197c 28.5 4.1
Hs12711662 28.5 4.6
CE18761 28.1 5.9
CE05993 28.1 6.4
Hs20562143 27.7 7.3
Hs14720380 27.7 7.5
Hs20373179 27.7 8.4
> SPBC119.18
Length=69
Score = 55.1 bits (131), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 22/51 (43%), Positives = 35/51 (68%), Gaps = 1/51 (1%)
Query 95 CAGLKEQYDRCFNHWYRHHFLQGDM-SRNCFHFFADYRVCISEELKQKGLE 144
C K++YD CFN WY + FL+GD+ +R+C FA+Y+ C+ + LK K ++
Sbjct 9 CTPAKKKYDACFNDWYANKFLKGDLHNRDCDELFAEYKSCLLKALKTKKID 59
> Hs7705467
Length=76
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Query 88 MREAMASCAGLKEQYDRCFNHWYRHHFLQGDMSRN-CFHFFADYRVCISEELKQKGL 143
M +C +K +YD+CFN W+ FL+GD S + C F Y+ C+ + +K+K +
Sbjct 1 MNSVGEACTDMKREYDQCFNRWFAEKFLKGDSSGDPCTDLFKRYQQCVQKAIKEKEI 57
> At4g33100
Length=92
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 31/63 (49%), Gaps = 1/63 (1%)
Query 82 KDHGIAMREAMASCAGLKEQYDRCFNHWYRHHFLQGDMSR-NCFHFFADYRVCISEELKQ 140
K + R + + CA L+ Y CFN WY F++G + C + YR C+SE L
Sbjct 6 KKDSTSARSSTSPCADLRNAYHNCFNKWYSEKFVKGQWDKEECVAEWKKYRDCLSENLDG 65
Query 141 KGL 143
K L
Sbjct 66 KLL 68
> YKL053c-a
Length=86
Score = 44.7 bits (104), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Query 95 CAGLKEQYDRCFNHWYRHHFLQGDMSRN-CFHFFADYRVCISEELKQKGLE 144
C LK +YD CFN WY FL+G N C + Y C++ L ++G++
Sbjct 13 CTDLKTKYDSCFNEWYSEKFLKGKSVENECSKQWYAYTTCVNAALVKQGIK 63
> YDR031w
Length=117
Score = 31.6 bits (70), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 14/58 (24%), Positives = 29/58 (50%), Gaps = 4/58 (6%)
Query 87 AMREAMASCAGLKEQYDRCFNHWYRHHFLQGDMSRNCFHFFADYRVCISEELKQKGLE 144
+++ M+ C+ ++YD+C R + ++ NC F D R C ++K K ++
Sbjct 55 SVKSIMSECSEPMKKYDQCI----RDNMGTRTINENCLGFLQDLRKCAELQVKNKNIK 108
> Hs18575668
Length=91
Score = 30.8 bits (68), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 5/49 (10%)
Query 100 EQYDRCFNHWYRHHFLQGDMSRN-CFHFFADYRVCI----SEELKQKGL 143
+ YD+CF+ W+ L+GD S C + F Y C+ +E + +GL
Sbjct 14 QSYDQCFSCWFVEKLLKGDSSGEPCSNLFKHYLQCVQKATTERISVEGL 62
> Hs18375615
Length=553
Score = 29.6 bits (65), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 29/75 (38%), Gaps = 17/75 (22%)
Query 8 GALAATQQAAGLVAEAWAVVVAINEGASFFLTQTLMP--------SLTPSVTAGPEAAST 59
G ATQQ W V A+F TQT+MP LTP T S
Sbjct 343 GLFPATQQP-------WPTVAGQFPPAAFMPTQTVMPLPAAMFQGPLTPLATV--PGTSD 393
Query 60 SSRSNSTCSAPGQAM 74
S+RS+ P Q M
Sbjct 394 STRSSPQTDKPRQKM 408
> Hs22045321
Length=426
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 33/73 (45%), Gaps = 7/73 (9%)
Query 8 GALAATQQAAGLVAEAWAVVVAINEGASFFLTQTLMPSLTPSVTAGPEAASTSSRSNSTC 67
G LA AG+ A+ V + ++ +F +L P+L P T PEA S + C
Sbjct 121 GNLALCDLLAGI---AYKVNILMSGKKTF----SLSPTLHPKQTNAPEADLKGSEAEKLC 173
Query 68 SAPGQAMPAATQQ 80
G++ A T Q
Sbjct 174 EGRGESGMAGTGQ 186
> 7303166
Length=3257
Score = 29.3 bits (64), Expect = 2.6, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 54 PEAASTSSRSNSTCSAPGQAMPAATQQPKDHGIAMREAMASC 95
PE T R + + S P + P +Q K+ G M E A+C
Sbjct 469 PEPLGTQLRRSKSESLPKRGRPRGQKQRKNRGATMEEKSANC 510
> Hs6912392
Length=558
Score = 28.9 bits (63), Expect = 3.1, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 23/45 (51%), Gaps = 5/45 (11%)
Query 37 FLTQTLMPSLTPSVTAGPEAASTSSRSNSTCSAPGQAMPAATQQP 81
+L + L+PS AGP A +++ ++ G+ PAA QP
Sbjct 4 YLAKALLPS-----RAGPAALGSAANHSAALLGRGRGQPAAASQP 43
> 7302280
Length=156
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 22/45 (48%), Gaps = 3/45 (6%)
Query 91 AMASCAGLKEQYDRC--FNHWYRHHFLQGDMSRNCFHFFADYRVC 133
A+ C KE+YD C F + +F+ G +C + DYR C
Sbjct 24 AIRPCHLYKEEYDDCTSFKARFHQYFIFG-KDTDCSQWLTDYRNC 67
> YNL197c
Length=661
Score = 28.5 bits (62), Expect = 4.1, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 30/58 (51%), Gaps = 0/58 (0%)
Query 24 WAVVVAINEGASFFLTQTLMPSLTPSVTAGPEAASTSSRSNSTCSAPGQAMPAATQQP 81
W A G+SFFL ++ P+ + A+ SS + ++ + QA+ A++QQP
Sbjct 355 WGNTSASQHGSSFFLPSAASTAIAPTNSNTSANANASSNNGASNNGANQALSASSQQP 412
> Hs12711662
Length=314
Score = 28.5 bits (62), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 90 EAMASCAGLKEQYDRCFNHWYRHHFL 115
AMAS A +QY+R F+ Y H++L
Sbjct 138 RAMASVARTPQQYERSFHFKYLHYYL 163
> CE18761
Length=1087
Score = 28.1 bits (61), Expect = 5.9, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Query 44 PSLTPSVTAGPEAA---STSSRSNSTCSAPGQAMPAATQQPKDHGIAMREA 91
PS + S AGP A S +S S ST + P +A PA +P +A + A
Sbjct 1008 PSRSNSNVAGPRRALELSGNSSSASTPAGPSKAQPAVV-RPSSKSVASKPA 1057
> CE05993
Length=1250
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Query 44 PSLTPSVTAGPEAA---STSSRSNSTCSAPGQAMPAATQQPKDHGIAMREA 91
PS + S AGP A S +S S ST + P +A PA +P +A + A
Sbjct 1048 PSRSNSNVAGPRRALELSGNSSSASTPAGPSKAQPAVV-RPSSKSVASKPA 1097
> Hs20562143
Length=583
Score = 27.7 bits (60), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 29/64 (45%), Gaps = 8/64 (12%)
Query 38 LTQTLMPSLTPSVTAGPEAASTSSRSNSTCSAPGQAMPAATQQPKDHGIAMRE---AMAS 94
L T PSL S+ PE S S+ +C AP + P + G+++ E A+A
Sbjct 147 LMDTYSPSLPKSLYRSPEKYEDLSSSDESCPAPQRQRPC-----RKKGVSIHEGPRALAR 201
Query 95 CAGL 98
G+
Sbjct 202 ITGI 205
> Hs14720380
Length=140
Score = 27.7 bits (60), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 7/51 (13%)
Query 52 AGPEAASTSSRSNSTCSAPGQAMPAATQQPKDHGIAMREAMASCAGLKEQY 102
AG AA +S+R PG P+A P+ G+ + +A A C L +Q+
Sbjct 17 AGLCAAGSSTR-------PGSMTPSANALPRSTGLTLPQACAFCDSLPQQF 60
> Hs20373179
Length=393
Score = 27.7 bits (60), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 14 QQAAGLVAEAWAVVVAINEGASFFLTQTLM 43
Q A GL+A W V + I +++F T+T++
Sbjct 177 QTATGLIALVWTVSILIAIPSAYFTTETVL 206
Lambda K H
0.318 0.126 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1675978996
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40