bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
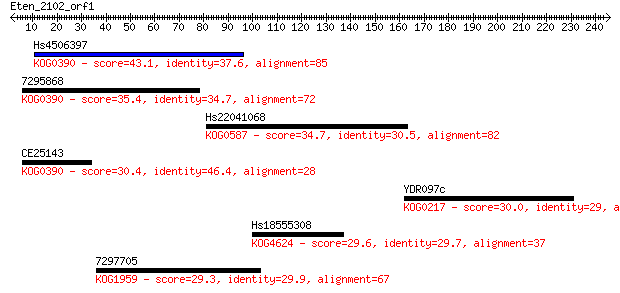
Query= Eten_2102_orf1
Length=245
Score E
Sequences producing significant alignments: (Bits) Value
Hs4506397 43.1 5e-04
7295868 35.4 0.12
Hs22041068 34.7 0.20
CE25143 30.4 3.4
YDR097c 30.0 4.0
Hs18555308 29.6 5.3
7297705 29.3 7.5
> Hs4506397
Length=747
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 43/87 (49%), Gaps = 8/87 (9%)
Query 11 VKDLFQLRE-TRSDTHDMLQCQRCQPSRETEPLGMVAQLVDGFE-EDDLLTWAHHSDLKT 68
+K+LF L E + SDTHD L C+RC SR+ P DG + DL W H +D
Sbjct 661 LKELFILDEASLSDTHDRLHCRRCVNSRQIRPPP------DGSDCTSDLAGWNHCTDKWG 714
Query 69 VPDEMLLKAIEAAEGDAADALSSDTHE 95
+ DE+L A +AA +HE
Sbjct 715 LRDEVLQAAWDAASTAITFVFHQHSHE 741
> 7295868
Length=784
Score = 35.4 bits (80), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 38/77 (49%), Gaps = 14/77 (18%)
Query 6 LSTDIVKDLFQL-RETRSDTHDMLQCQRCQPSRETEPLGMVAQLVDGFEEDD----LLTW 60
+ D +KDLF SDTHD L+C+RC + + +P E+ D L W
Sbjct 659 FTRDDLKDLFTFDANILSDTHDKLKCKRCVQNIQMKPPP---------EDTDCTSHLSQW 709
Query 61 AHHSDLKTVPDEMLLKA 77
H S+ + +PD +L +A
Sbjct 710 YHCSNNRGLPDNILAQA 726
> Hs22041068
Length=810
Score = 34.7 bits (78), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 40/82 (48%), Gaps = 2/82 (2%)
Query 81 AEGDAADALSSDTHEAPTEIPSHFQPVSFAMSCKVEFTKESEETNSAGAPAPKKVNHGEA 140
++G A A S HE PT+ S FQ S +VE +++ + S P P ++ +
Sbjct 50 SQGPALTA-SQSVHEQPTKGLSGFQEALNVTSHRVEMPRQNSDPTSENPPLPTRIEKFDR 108
Query 141 CLSEMRVVSSAPPTSPQQTTAL 162
S +R PP PQ+TT++
Sbjct 109 S-SWLRQEEDIPPKVPQRTTSI 129
> CE25143
Length=818
Score = 30.4 bits (67), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 22/29 (75%), Gaps = 1/29 (3%)
Query 6 LSTDIVKDLFQLRET-RSDTHDMLQCQRC 33
S++ +++LF+L T SDTH+ L+C+RC
Sbjct 707 FSSEQLRELFKLESTVASDTHEKLKCKRC 735
> YDR097c
Length=1242
Score = 30.0 bits (66), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 20/74 (27%), Positives = 33/74 (44%), Gaps = 5/74 (6%)
Query 162 LGQYSSLENALVKPIDR-----GERPGILQNRENVCVNTSVTTACAEAPSREALHAVDDT 216
+G Y E+A++ PIDR G I+Q + V + T + + +L VD+
Sbjct 1004 MGCYVPCESAVLTPIDRIMTRLGANDNIMQGKSTFFVELAETKKILDMATNRSLLVVDEL 1063
Query 217 SDVSCSSEGTITSE 230
SS+G +E
Sbjct 1064 GRGGSSSDGFAIAE 1077
> Hs18555308
Length=106
Score = 29.6 bits (65), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 11/37 (29%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 100 IPSHFQPVSFAMSCKVEFTKESEETNSAGAPAPKKVN 136
+ +++ +F CK+E+ KE EE G P K++
Sbjct 64 LTAYYNDPAFYEECKMEYLKEREEFRKTGIPTKKRLQ 100
> 7297705
Length=950
Score = 29.3 bits (64), Expect = 7.5, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 32/67 (47%), Gaps = 0/67 (0%)
Query 36 SRETEPLGMVAQLVDGFEEDDLLTWAHHSDLKTVPDEMLLKAIEAAEGDAADALSSDTHE 95
++ +E L ++Q + + D +T + + D +L KAI AE A DAL S T+
Sbjct 360 TQYSEDLDNLSQAMGIMQHHDAVTGTEKQAVASDYDRLLFKAIVGAENSARDALRSLTNL 419
Query 96 APTEIPS 102
E S
Sbjct 420 TSGEFES 426
Lambda K H
0.308 0.124 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4938804274
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40