bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2090_orf3
Length=414
Score E
Sequences producing significant alignments: (Bits) Value
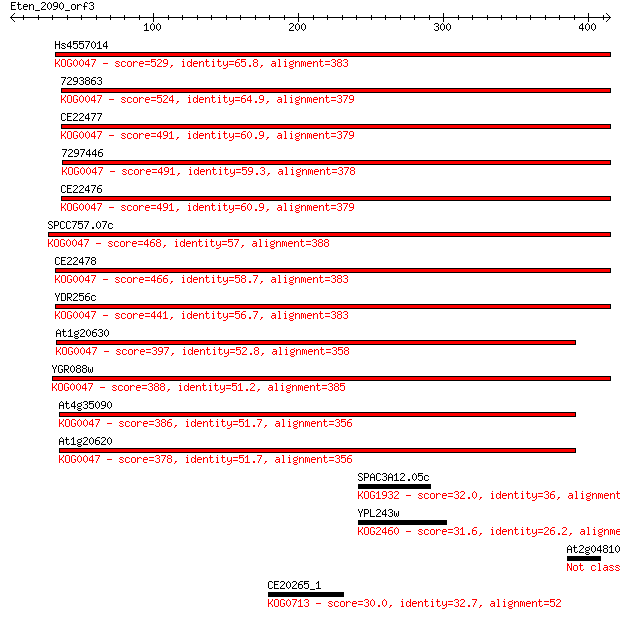
Hs4557014 529 4e-150
7293863 524 1e-148
CE22477 491 1e-138
7297446 491 1e-138
CE22476 491 1e-138
SPCC757.07c 468 1e-131
CE22478 466 5e-131
YDR256c 441 2e-123
At1g20630 397 3e-110
YGR088w 388 1e-107
At4g35090 386 4e-107
At1g20620 378 1e-104
SPAC3A12.05c 32.0 2.5
YPL243w 31.6 3.0
At2g04810 30.4 8.0
CE20265_1 30.0 9.9
> Hs4557014
Length=527
Score = 529 bits (1363), Expect = 4e-150, Method: Compositional matrix adjust.
Identities = 252/386 (65%), Positives = 295/386 (76%), Gaps = 6/386 (1%)
Query 32 KECPVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGA 91
++ VLTT G PVGD N +T GP G +L+ D D++AHFDRERIPERVVHAKGAGA
Sbjct 22 QKADVLTTGAGNPVGDKLNVITVGPRGPLLVQDVVFTDEMAHFDRERIPERVVHAKGAGA 81
Query 92 HGVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAG 151
G F+VTHDITKY KA+VFE +GK+T + VRFSTV GE GSAD+ RDPRGFAVKFY+ G
Sbjct 82 FGYFEVTHDITKYSKAKVFEHIGKKTPIAVRFSTVAGESGSADTVRDPRGFAVKFYTEDG 141
Query 152 VWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFS 211
WDLVGNNTP+FFIRDP+ FP FIHSQKRNPQT+L DP+M WDF SL PE+LHQ + LFS
Sbjct 142 NWDLVGNNTPIFFIRDPILFPSFIHSQKRNPQTHLKDPDMVWDFWSLRPESLHQVSFLFS 201
Query 212 DRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYA 271
DRG PDG+RHM+GY SH FKLVN+ G Y KFH KT+Q IKNLS + A +L+ EDPDY
Sbjct 202 DRGIPDGHRHMNGYGSHTFKLVNANGEAVYCKFHYKTDQGIKNLSVEDAARLSQEDPDYG 261
Query 272 TRDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKN 331
RDLFNAI +G +PSWT YIQVM +A + +N D+TKV P KD+PL+PVG+L L +N
Sbjct 262 IRDLFNAIATGKYPSWTFYIQVMTFNQAETFPFNPFDLTKVWPHKDYPLIPVGKLVLNRN 321
Query 332 PKNYFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCP 391
P NYF EVEQ+AF P N+ PGIE S DK+LQGRLFAYPDT RHRLGPN+ IPVN CP
Sbjct 322 PVNYFAEVEQIAFDPSNMPPGIEASPDKMLQGRLFAYPDTHRHRLGPNYLHIPVN---CP 378
Query 392 LRAI---HIRDGPMCVNGNYGALPNY 414
RA + RDGPMC+ N G PNY
Sbjct 379 YRARVANYQRDGPMCMQDNQGGAPNY 404
> 7293863
Length=506
Score = 524 bits (1350), Expect = 1e-148, Method: Compositional matrix adjust.
Identities = 246/379 (64%), Positives = 294/379 (77%), Gaps = 0/379 (0%)
Query 36 VLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGVF 95
+TT GAP+G S T GP G +LL D + LD+++HFDRERIPERVVHAKGAGA G F
Sbjct 24 AITTGNGAPIGIKDASQTVGPRGPILLQDVNFLDEMSHFDRERIPERVVHAKGAGAFGYF 83
Query 96 KVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWDL 155
+VTHDIT+YC A++F+ + K T + VRFSTVGGE GSAD+ARDPRGFAVKFY+ GVWDL
Sbjct 84 EVTHDITQYCAAKIFDKVKKRTPLAVRFSTVGGESGSADTARDPRGFAVKFYTEDGVWDL 143
Query 156 VGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRGT 215
VGNNTPVFFIRDP+ FP FIH+QKRNPQT+L DP+MFWDFL+L PE+ HQ ILFSDRGT
Sbjct 144 VGNNTPVFFIRDPILFPSFIHTQKRNPQTHLKDPDMFWDFLTLRPESAHQVCILFSDRGT 203
Query 216 PDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDL 275
PDGY HM+GY SH FKL+N++G YAKFH KT+Q IKNL + A QLA DPDY+ RDL
Sbjct 204 PDGYCHMNGYGSHTFKLINAKGEPIYAKFHFKTDQGIKNLDVKTADQLASTDPDYSIRDL 263
Query 276 FNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKNY 335
+N I++ FPSWT+YIQVM +A K+KYN DVTKV QK++PL+PVG++ L +NPKNY
Sbjct 264 YNRIKTCKFPSWTMYIQVMTYEQAKKFKYNPFDVTKVWSQKEYPLIPVGKMVLDRNPKNY 323
Query 336 FQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCPLRAI 395
F EVEQ+AFSP +L+PG+EPS DK+L GRLF+Y DT RHRLGPN+ QIPVN P
Sbjct 324 FAEVEQIAFSPAHLVPGVEPSPDKMLHGRLFSYSDTHRHRLGPNYLQIPVNCPYKVKIEN 383
Query 396 HIRDGPMCVNGNYGALPNY 414
RDG M V N PNY
Sbjct 384 FQRDGAMNVTDNQDGAPNY 402
> CE22477
Length=524
Score = 491 bits (1265), Expect = 1e-138, Method: Compositional matrix adjust.
Identities = 231/382 (60%), Positives = 288/382 (75%), Gaps = 7/382 (1%)
Query 36 VLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGVF 95
V+TTS GAP+ LTAG G +L+ D +D++AHFDRERIPERVVHAKGAGAHG F
Sbjct 46 VITTSNGAPIYSKTAVLTAGRRGPMLMQDVVYMDEMAHFDRERIPERVVHAKGAGAHGYF 105
Query 96 KVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWDL 155
+VTHDI+KYCKA++F +GK+T + +RFSTVGGE GSAD+ARDPRGFA+KFY+ G WDL
Sbjct 106 EVTHDISKYCKADIFNKVGKQTPLLIRFSTVGGESGSADTARDPRGFAIKFYTEEGNWDL 165
Query 156 VGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRGT 215
VGNNTP+FFIRDP+ FP+FIH+QKRNPQT+L DPNM +DF PE LHQ LFSDRG
Sbjct 166 VGNNTPIFFIRDPIHFPNFIHTQKRNPQTHLKDPNMIFDFWLHRPEALHQVMFLFSDRGL 225
Query 216 PDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDL 275
PDGYRHM+GY SH FK+VN +G Y KFH K Q +KNL+ ++A QLA DPDY+ RDL
Sbjct 226 PDGYRHMNGYGSHTFKMVNKDGKAIYVKFHFKPTQGVKNLTVEKAGQLASSDPDYSIRDL 285
Query 276 FNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKNY 335
FNAI+ G+FP W ++IQVM +A K+++N DVTKV P D+PL+ VG++ L +NP+NY
Sbjct 286 FNAIEKGDFPVWKMFIQVMTFEQAEKWEFNPFDVTKVWPHGDYPLIEVGKMVLNRNPRNY 345
Query 336 FQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCPLRAI 395
F EVEQ AF P +++PGIE S DK+LQGR+F+Y DT HRLGPN+ Q+PVN CP R+
Sbjct 346 FAEVEQSAFCPAHIVPGIEFSPDKMLQGRIFSYTDTHFHRLGPNYIQLPVN---CPYRSR 402
Query 396 H---IRDGPMCVNGNYGALPNY 414
RDG M + A PN+
Sbjct 403 AHNTQRDGAMAYDNQQHA-PNF 423
> 7297446
Length=506
Score = 491 bits (1264), Expect = 1e-138, Method: Compositional matrix adjust.
Identities = 224/379 (59%), Positives = 282/379 (74%), Gaps = 2/379 (0%)
Query 37 LTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGVFK 96
+TTS+G PVG T GP G LL DF LD++ HFD ERIPERV +AKGAGA G F+
Sbjct 25 ITTSSGTPVGVKDAIQTVGPRGPALLQDFQFLDEVMHFDSERIPERVAYAKGAGAFGYFE 84
Query 97 VTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWDLV 156
THDI+K+C A +F+ + K T V +RFS GE+GSAD+ R+ RGFAVKFY+ G+WD+V
Sbjct 85 CTHDISKFCAASIFDKVRKRTAVAMRFSVACGEQGSADTVREQRGFAVKFYTDDGIWDIV 144
Query 157 GNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRGTP 216
G N PV ++RDP+ FP +H+QKRNPQT+L DP+MFWDF++L PETLH + FSDRGTP
Sbjct 145 GCNMPVHYVRDPMLFPSLVHAQKRNPQTHLKDPDMFWDFMTLRPETLHALLMYFSDRGTP 204
Query 217 DGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDLF 276
DGYRH+HGY H ++++N+ G Y +FH KT+Q IKNL +R ++L DPDYA RDL+
Sbjct 205 DGYRHLHGYGVHTYRMINASGETQYVRFHFKTDQGIKNLDARRCEELMSHDPDYAIRDLY 264
Query 277 NAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKNYF 336
N+I+ GN+PSW++YIQVM EA K ++N DVTKV PQKDFPLLPVG++ L +NP NYF
Sbjct 265 NSIKKGNYPSWSMYIQVMLNEEAKKCRFNPFDVTKVWPQKDFPLLPVGKIVLDRNPTNYF 324
Query 337 QEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRP-RCPLRAI 395
EVEQ+AFSP +++PGIEPS DK+LQGRLFAY D+QRHRLG N+ QIPVN P R +R
Sbjct 325 TEVEQLAFSPAHMVPGIEPSPDKMLQGRLFAYGDSQRHRLGVNYMQIPVNCPYRVNVRNF 384
Query 396 HIRDGPMCVNGNYGALPNY 414
RDG M V N PNY
Sbjct 385 Q-RDGAMTVTDNQNGAPNY 402
> CE22476
Length=500
Score = 491 bits (1264), Expect = 1e-138, Method: Compositional matrix adjust.
Identities = 231/382 (60%), Positives = 288/382 (75%), Gaps = 7/382 (1%)
Query 36 VLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGVF 95
V+TTS GAP+ LTAG G +L+ D +D++AHFDRERIPERVVHAKGAGAHG F
Sbjct 22 VITTSNGAPIYSKTAVLTAGRRGPMLMQDVVYMDEMAHFDRERIPERVVHAKGAGAHGYF 81
Query 96 KVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWDL 155
+VTHDI+KYCKA++F +GK+T + +RFSTVGGE GSAD+ARDPRGFA+KFY+ G WDL
Sbjct 82 EVTHDISKYCKADIFNKVGKQTPLLIRFSTVGGESGSADTARDPRGFAIKFYTEEGNWDL 141
Query 156 VGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRGT 215
VGNNTP+FFIRDP+ FP+FIH+QKRNPQT+L DPNM +DF PE LHQ LFSDRG
Sbjct 142 VGNNTPIFFIRDPIHFPNFIHTQKRNPQTHLKDPNMIFDFWLHRPEALHQVMFLFSDRGL 201
Query 216 PDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDL 275
PDGYRHM+GY SH FK+VN +G Y KFH K Q +KNL+ ++A QLA DPDY+ RDL
Sbjct 202 PDGYRHMNGYGSHTFKMVNKDGKAIYVKFHFKPTQGVKNLTVEKAGQLASSDPDYSIRDL 261
Query 276 FNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKNY 335
FNAI+ G+FP W ++IQVM +A K+++N DVTKV P D+PL+ VG++ L +NP+NY
Sbjct 262 FNAIEKGDFPVWKMFIQVMTFEQAEKWEFNPFDVTKVWPHGDYPLIEVGKMVLNRNPRNY 321
Query 336 FQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCPLRAI 395
F EVEQ AF P +++PGIE S DK+LQGR+F+Y DT HRLGPN+ Q+PVN CP R+
Sbjct 322 FAEVEQSAFCPAHIVPGIEFSPDKMLQGRIFSYTDTHFHRLGPNYIQLPVN---CPYRSR 378
Query 396 H---IRDGPMCVNGNYGALPNY 414
RDG M + A PN+
Sbjct 379 AHNTQRDGAMAYDNQQHA-PNF 399
> SPCC757.07c
Length=512
Score = 468 bits (1204), Expect = 1e-131, Method: Compositional matrix adjust.
Identities = 221/389 (56%), Positives = 283/389 (72%), Gaps = 2/389 (0%)
Query 27 STMEEKECPVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHA 86
++ + PV TT+TG P+ + + G G VLL D HL+D HFDRERIPERVVHA
Sbjct 2 NSKDSNTVPVYTTNTGCPIFNPMAAARVGKGGPVLLQDSHLIDVFQHFDRERIPERVVHA 61
Query 87 KGAGAHGVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKF 146
KG+GA G F+ T DITKY K +F +GK+T + RFSTVGGE+G+ D+ARDPRGFA+KF
Sbjct 62 KGSGAFGEFECTDDITKYTKHTMFSKVGKKTPMVARFSTVGGERGTPDTARDPRGFALKF 121
Query 147 YSAAGVWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQT 206
Y+ G++D+VGNNTPVFF+RDP KFP FIH+QKRNPQ ++ D MFWD+LS + E++HQ
Sbjct 122 YTDEGIFDMVGNNTPVFFLRDPAKFPLFIHTQKRNPQNDMKDATMFWDYLSQNAESIHQV 181
Query 207 TILFSDR-GTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAG 265
ILFSD GTP YR M G+SSH +K VN +G +Y K+H TNQ K L+ + A L G
Sbjct 182 MILFSDLGGTPYSYRFMDGFSSHTYKFVNDKGEFYYCKWHFITNQGTKGLTNEEAAALDG 241
Query 266 EDPDYATRDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQ 325
+PD+A +DLF AI+ G++PSWTLY+QVM EA KY+YN+ D+TKV P KD P+ VG+
Sbjct 242 SNPDHARQDLFEAIERGDYPSWTLYVQVMTPQEAEKYRYNIFDLTKVWPHKDVPMQRVGR 301
Query 326 LTLTKNPKNYFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPV 385
TL +NP N+F ++EQ FSP +++PGIE S D +LQ R F+YPDT RHRLG NF+QIPV
Sbjct 302 FTLNQNPTNFFADIEQAGFSPSHMVPGIEVSADPVLQVRTFSYPDTHRHRLGANFEQIPV 361
Query 386 NRPRCPLRAIHIRDGPMCVNGNYGALPNY 414
N P+CP+ + RDGPM VNGN G PNY
Sbjct 362 NSPKCPVFN-YSRDGPMNVNGNQGNWPNY 389
> CE22478
Length=512
Score = 466 bits (1198), Expect = 5e-131, Method: Compositional matrix adjust.
Identities = 225/386 (58%), Positives = 279/386 (72%), Gaps = 7/386 (1%)
Query 32 KECPVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGA 91
+E +LTTS GAP+ LTAG G +L+ D +D++AHFDRERIPERVVHAKG GA
Sbjct 33 QEPHLLTTSNGAPIYSKTAVLTAGRRGPMLMQDIVYMDEMAHFDRERIPERVVHAKGGGA 92
Query 92 HGVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAG 151
HG F+VTHDITKYCKA++F +GK+T + VRFSTV GE GSAD+ RDPRGF++KFY+ G
Sbjct 93 HGYFEVTHDITKYCKADMFNKVGKQTPLLVRFSTVAGESGSADTVRDPRGFSLKFYTEEG 152
Query 152 VWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFS 211
WDLVGNNTP+FFIRD + FP+FIH+ KRNPQT++ DPN +DF PE++HQ L+S
Sbjct 153 NWDLVGNNTPIFFIRDAIHFPNFIHALKRNPQTHMRDPNALFDFWMNRPESIHQVMFLYS 212
Query 212 DRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYA 271
DRG PDG+R M+GY +H FK+VN EG Y KFH K Q KNL A +LA DPDYA
Sbjct 213 DRGIPDGFRFMNGYGAHTFKMVNKEGNPIYCKFHFKPAQGSKNLDPTDAGKLASSDPDYA 272
Query 272 TRDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKN 331
RDLFNAI+S NFP W ++IQVM +A K+++N DVTKV P D+PL+ VG++ L +N
Sbjct 273 IRDLFNAIESRNFPEWKMFIQVMTFEQAEKWEFNPFDVTKVWPHGDYPLIEVGKMVLNRN 332
Query 332 PKNYFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCP 391
KNYF EVEQ AF P +++PGIE S DK+LQGR+F+Y DT HRLGPN+ Q+PVN CP
Sbjct 333 VKNYFAEVEQAAFCPAHIVPGIEFSPDKMLQGRIFSYTDTHYHRLGPNYIQLPVN---CP 389
Query 392 LRA-IHI--RDGPMCVNGNYGALPNY 414
R+ H RDG M G PNY
Sbjct 390 YRSRAHTTQRDGAMAYESQ-GDAPNY 414
> YDR256c
Length=515
Score = 441 bits (1133), Expect = 2e-123, Method: Compositional matrix adjust.
Identities = 217/390 (55%), Positives = 273/390 (70%), Gaps = 11/390 (2%)
Query 32 KECPVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGA 91
+E V+T STG P+ + + G +G +LL D++L+D LAHF+RE IP+R HA G+GA
Sbjct 17 REDRVVTNSTGNPINEPFVTQRIGEHGPLLLQDYNLIDSLAHFNRENIPQRNPHAHGSGA 76
Query 92 HGVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAG 151
G F+VT DIT C + +F +GK T+ RFSTVGG+KGSAD+ RDPRGFA KFY+ G
Sbjct 77 FGYFEVTDDITDICGSAMFSKIGKRTKCLTRFSTVGGDKGSADTVRDPRGFATKFYTEEG 136
Query 152 VWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPE---TLHQTTI 208
D V NNTPVFFIRDP KFP FIH+QKRNPQTNL D +MFWDFL+ +PE +HQ I
Sbjct 137 NLDWVYNNTPVFFIRDPSKFPHFIHTQKRNPQTNLRDADMFWDFLT-TPENQVAIHQVMI 195
Query 209 LFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDP 268
LFSDRGTP YR MHGYS H +K N G HY + H+KT+Q IKNL+ + A ++AG +P
Sbjct 196 LFSDRGTPANYRSMHGYSGHTYKWSNKNGDWHYVQVHIKTDQGIKNLTIEEATKIAGSNP 255
Query 269 DYATRDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTL 328
DY +DLF AIQ+GN+PSWT+YIQ M +A K ++V D+TKV PQ FPL VG++ L
Sbjct 256 DYCQQDLFEAIQNGNYPSWTVYIQTMTERDAKKLPFSVFDLTKVWPQGQFPLRRVGKIVL 315
Query 329 TKNPKNYFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRP 388
+NP N+F +VEQ AF+P +P E S D +LQ RLF+Y D R+RLGPNF QIPVN
Sbjct 316 NENPLNFFAQVEQAAFAPSTTVPYQEASADPVLQARLFSYADAHRYRLGPNFHQIPVN-- 373
Query 389 RCPLRAIH----IRDGPMCVNGNYGALPNY 414
CP + IRDGPM VNGN+G+ P Y
Sbjct 374 -CPYASKFFNPAIRDGPMNVNGNFGSEPTY 402
> At1g20630
Length=492
Score = 397 bits (1019), Expect = 3e-110, Method: Compositional matrix adjust.
Identities = 189/358 (52%), Positives = 252/358 (70%), Gaps = 0/358 (0%)
Query 33 ECPVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAH 92
+ P TT++GAPV +N +SLT G G +LL D+HLL+KLA+FDRERIPERVVHA+GA A
Sbjct 13 DSPFFTTNSGAPVWNNNSSLTVGTRGPILLEDYHLLEKLANFDRERIPERVVHARGASAK 72
Query 93 GVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGV 152
G F+VTHDIT+ A+ G G +T V VRFSTV E+GS ++ RDPRGFAVKFY+ G
Sbjct 73 GFFEVTHDITQLTSADFLRGPGVQTPVIVRFSTVIHERGSPETLRDPRGFAVKFYTREGN 132
Query 153 WDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSD 212
+DLVGNN PVFF+RD +KFPD +H+ K NP++++ + DF S PE+LH + LF D
Sbjct 133 FDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHPESLHMFSFLFDD 192
Query 213 RGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYAT 272
G P YRHM G + + L+N G HY KFH K IK LS + A ++ G + +AT
Sbjct 193 LGIPQDYRHMEGAGVNTYMLINKAGKAHYVKFHWKPTCGIKCLSDEEAIRVGGANHSHAT 252
Query 273 RDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNP 332
+DL+++I +GN+P W L++QVM A K+ ++ LDVTK+ P+ PL PVG+L L KN
Sbjct 253 KDLYDSIAAGNYPQWNLFVQVMDPAHEDKFDFDPLDVTKIWPEDILPLQPVGRLVLNKNI 312
Query 333 KNYFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRC 390
N+F E EQ+AF P ++PGI S+DKLLQ R+F+Y D+QRHRLGPN+ Q+PVN P+C
Sbjct 313 DNFFNENEQIAFCPALVVPGIHYSDDKLLQTRIFSYADSQRHRLGPNYLQLPVNAPKC 370
> YGR088w
Length=573
Score = 388 bits (996), Expect = 1e-107, Method: Compositional matrix adjust.
Identities = 197/408 (48%), Positives = 266/408 (65%), Gaps = 24/408 (5%)
Query 30 EEKECPVLTTSTGAPVGDN-QNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKG 88
EEK+ V + G P + S + P G +LL DFHLL+ +A FDRER+PERVVHAKG
Sbjct 19 EEKQEKVYSLQNGFPYSHHPYASQYSRPDGPILLQDFHLLENIASFDRERVPERVVHAKG 78
Query 89 AGAHGVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYS 148
G F++T ++ A ++ +G + VRFSTVGGE G+ D+ARDPRG + KFY+
Sbjct 79 GGCRLEFELTDSLSDITYAAPYQNVGYKCPGLVRFSTVGGESGTPDTARDPRGVSFKFYT 138
Query 149 AAGVWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNL---PDPNMFWDFLSLSPETLHQ 205
G D V NNTPVFF+RD +KFP FIHSQKR+PQ++L D ++WD+L+L+PE++HQ
Sbjct 139 EWGNHDWVFNNTPVFFLRDAIKFPVFIHSQKRDPQSHLNQFQDTTIYWDYLTLNPESIHQ 198
Query 206 TTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAG 265
T +F DRGTP + M+ YS H+F +VN EG Y +FH+ ++ + L+ +A +L+G
Sbjct 199 ITYMFGDRGTPASWASMNAYSGHSFIMVNKEGKDTYVQFHVLSDTGFETLTGDKAAELSG 258
Query 266 EDPDYATRDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQ 325
PDY LF +Q+G P + Y+Q M +A K++Y+V D+TK+ P K+FPL G
Sbjct 259 SHPDYNQAKLFTQLQNGEKPKFNCYVQTMTPEQATKFRYSVNDLTKIWPHKEFPLRKFGT 318
Query 326 LTLTKNPKNYFQEVEQVAFSPGN-LIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIP 384
+TLT+N NYFQE+EQVAFSP N IPGI+PS D +LQ RLF+YPDTQRHRLG N+ Q+P
Sbjct 319 ITLTENVDNYFQEIEQVAFSPTNTCIPGIKPSNDSVLQARLFSYPDTQRHRLGANYQQLP 378
Query 385 VNRPR-----------------CPLRAIHI-RDGPMCVNGNYGALPNY 414
VNRPR CP +A++ RDGPM N+G PNY
Sbjct 379 VNRPRNLGCPYSKGDSQYTAEQCPFKAVNFQRDGPMSYY-NFGPEPNY 425
> At4g35090
Length=492
Score = 386 bits (992), Expect = 4e-107, Method: Compositional matrix adjust.
Identities = 184/356 (51%), Positives = 250/356 (70%), Gaps = 0/356 (0%)
Query 35 PVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGV 94
P TT++GAPV +N +S+T GP G +LL D+HL++KLA+FDRERIPERVVHA+GA A G
Sbjct 15 PFFTTNSGAPVWNNNSSMTVGPRGPILLEDYHLVEKLANFDRERIPERVVHARGASAKGF 74
Query 95 FKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWD 154
F+VTHDI+ A+ G +T V VRFSTV E+GS ++ RDPRGFAVKFY+ G +D
Sbjct 75 FEVTHDISNLTCADFLRAPGVQTPVIVRFSTVIHERGSPETLRDPRGFAVKFYTREGNFD 134
Query 155 LVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRG 214
LVGNN PVFFIRD +KFPD +H+ K NP++++ + DF S PE+L+ T LF D G
Sbjct 135 LVGNNFPVFFIRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHPESLNMFTFLFDDIG 194
Query 215 TPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRD 274
P YRHM G + + L+N G HY KFH K +K+L + A ++ G + +AT+D
Sbjct 195 IPQDYRHMDGSGVNTYMLINKAGKAHYVKFHWKPTCGVKSLLEEDAIRVGGTNHSHATQD 254
Query 275 LFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKN 334
L+++I +GN+P W L+IQ++ A+ K+ ++ LDVTK P+ PL PVG++ L KN N
Sbjct 255 LYDSIAAGNYPEWKLFIQIIDPADEDKFDFDPLDVTKTWPEDILPLQPVGRMVLNKNIDN 314
Query 335 YFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRC 390
+F E EQ+AF P ++PGI S+DKLLQ R+F+Y DTQRHRLGPN+ Q+PVN P+C
Sbjct 315 FFAENEQLAFCPAIIVPGIHYSDDKLLQTRVFSYADTQRHRLGPNYLQLPVNAPKC 370
> At1g20620
Length=531
Score = 378 bits (970), Expect = 1e-104, Method: Compositional matrix adjust.
Identities = 184/356 (51%), Positives = 245/356 (68%), Gaps = 0/356 (0%)
Query 35 PVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGV 94
P TT+ GAPV +N +SLT G G VLL D+HL++K+A+F RERIPERVVHA+G A G
Sbjct 54 PFYTTNGGAPVSNNISSLTIGERGPVLLEDYHLIEKVANFTRERIPERVVHARGISAKGF 113
Query 95 FKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWD 154
F+VTHDI+ A+ G +T V VRFSTV E+ S ++ RD RGFAVKFY+ G +D
Sbjct 114 FEVTHDISNLTCADFLRAPGVQTPVIVRFSTVVHERASPETMRDIRGFAVKFYTREGNFD 173
Query 155 LVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRG 214
LVGNNTPVFFIRD ++FPD +H+ K NP+TN+ + D++S PE+L +F D G
Sbjct 174 LVGNNTPVFFIRDGIQFPDVVHALKPNPKTNIQEYWRILDYMSHLPESLLTWCWMFDDVG 233
Query 215 TPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRD 274
P YRHM G+ H + L+ G V + KFH K IKNL+ + A+ + G + +AT+D
Sbjct 234 IPQDYRHMEGFGVHTYTLIAKSGKVLFVKFHWKPTCGIKNLTDEEAKVVGGANHSHATKD 293
Query 275 LFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKN 334
L +AI SGN+P W L+IQ M A+ K+ ++ LDVTK+ P+ PL PVG+L L + N
Sbjct 294 LHDAIASGNYPEWKLFIQTMDPADEDKFDFDPLDVTKIWPEDILPLQPVGRLVLNRTIDN 353
Query 335 YFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRC 390
+F E EQ+AF+PG ++PGI S+DKLLQ R+FAY DTQRHRLGPN+ Q+PVN P+C
Sbjct 354 FFNETEQLAFNPGLVVPGIYYSDDKLLQCRIFAYGDTQRHRLGPNYLQLPVNAPKC 409
> SPAC3A12.05c
Length=1174
Score = 32.0 bits (71), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 28/58 (48%), Gaps = 8/58 (13%)
Query 241 YAKFHLKTNQKIKNLSRQRAQQLA-------GEDPDYATRDLFNAIQSG-NFPSWTLY 290
+ K ++ N K K +SR R+ + G+D DY R L + +QS + W LY
Sbjct 627 FTKLDIQYNTKYKRISRNRSTKNVKRDGDHNGDDSDYVIRSLGDVLQSDEDIERWHLY 684
> YPL243w
Length=599
Score = 31.6 bits (70), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 35/61 (57%), Gaps = 1/61 (1%)
Query 241 YAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDLFNAIQSGNFPSWTLYIQVMPVAEAP 300
+AK+H K N+K+++L R R + + Y++++ + I S ++ + T I V+ + A
Sbjct 25 FAKYHAKLNKKLQHL-RSRCHLVTKDTKKYSSKNKYGEINSEDYDNKTKLIGVLILLHAE 83
Query 301 K 301
+
Sbjct 84 R 84
> At2g04810
Length=397
Score = 30.4 bits (67), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 14/23 (60%), Gaps = 0/23 (0%)
Query 385 VNRPRCPLRAIHIRDGPMCVNGN 407
V RP CPLR IH D P +GN
Sbjct 61 VKRPLCPLRIIHQGDSPTVGDGN 83
> CE20265_1
Length=114
Score = 30.0 bits (66), Expect = 9.9, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 27/59 (45%), Gaps = 7/59 (11%)
Query 179 KRNPQTNLPDPNMFWDFLS-------LSPETLHQTTILFSDRGTPDGYRHMHGYSSHAF 230
K++P N DPN +F+ L E L + F ++G DG++ + Y S F
Sbjct 46 KKHPDRNTDDPNAHDEFVKINKAYEVLKDENLRKKYDQFGEKGLEDGFQGGNNYQSWQF 104
Lambda K H
0.318 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10594808898
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40