bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2054_orf1
Length=201
Score E
Sequences producing significant alignments: (Bits) Value
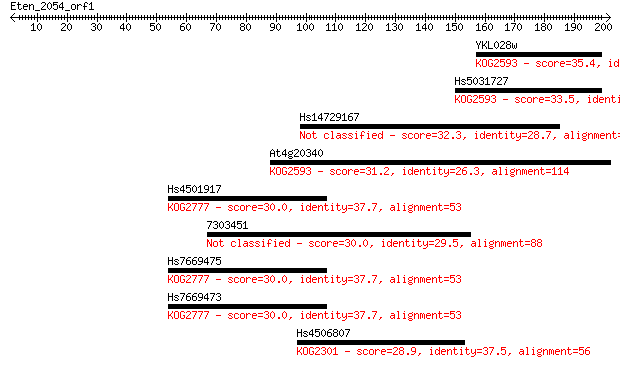
YKL028w 35.4 0.082
Hs5031727 33.5 0.28
Hs14729167 32.3 0.57
At4g20340 31.2 1.6
Hs4501917 30.0 3.3
7303451 30.0 3.3
Hs7669475 30.0 3.4
Hs7669473 30.0 3.5
Hs4506807 28.9 6.5
> YKL028w
Length=482
Score = 35.4 bits (80), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 11/42 (26%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 157 FYYRVSPYAVIVVQYRIEQIDTKLKQQRREAETRDVFVCPIC 198
+YY P+A+ +++++ Q+ +LK + + ++CPIC
Sbjct 86 YYYVKYPHAIDAIKWKVHQVVQRLKDDLDKNSEPNGYMCPIC 127
> Hs5031727
Length=439
Score = 33.5 bits (75), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 12/49 (24%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 150 GASSAAPFYYRVSPYAVIVVQYRIEQIDTKLKQQRREAETRDVFVCPIC 198
G ++ +Y+ V VV+Y+++ + +++ R++ R F CP+C
Sbjct 84 GKTTRHNYYFINYRTLVNVVKYKLDHMRRRIETDERDSTNRASFKCPVC 132
> Hs14729167
Length=1138
Score = 32.3 bits (72), Expect = 0.57, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 43/89 (48%), Gaps = 2/89 (2%)
Query 98 ERLMLIQKEQLSAASAQALSNASADGDTTSSVG--GTTDIGRGSMPGGAALLGSGASSAA 155
++ +L+ K++ A + + G VG TT + S+P G+ + SGAS A
Sbjct 667 QKRLLLMKQKGVMNQPMAYAALPSHGQEQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAG 726
Query 156 PFYYRVSPYAVIVVQYRIEQIDTKLKQQR 184
P + P A I+ Q I+Q ++QQ+
Sbjct 727 PGFLGSQPQAAIMKQMLIDQRAQLIEQQK 755
> At4g20340
Length=527
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 47/119 (39%), Gaps = 19/119 (15%)
Query 88 KRVREVCASLERLMLIQKEQLSAASAQALSNASADGDTTSSVGGTTDIGRGSMPGGAALL 147
K V+ L +L+ +EQ A + +VGGTTD GR
Sbjct 70 KEVKRNAKELRKLIRHFEEQKFVMRYHRKETAKRAKMYSYAVGGTTD-GR---------- 118
Query 148 GSGASSAAPFY---YRVSPYAVI--VVQYRIEQIDTKLKQQRREAETRDVFVCPICGRQ 201
A F+ Y YA I +V+Y++ ++ K K + + T + CP C R+
Sbjct 119 ---AEDNVKFHTHSYCCLDYAQIYDIVRYKLHRLKKKFKDELEDRNTVQEYGCPNCKRK 174
> Hs4501917
Length=1226
Score = 30.0 bits (66), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query 54 LFCDDEQIVVVDFLAS--EEKAFTERELIDRLGWPDKRVREVCASLERLMLIQKE 106
++ D EQ ++ FL E KA T +L +LG P K + V SL + +QKE
Sbjct 135 IYQDQEQ-RILKFLEELGEGKATTAHDLSGKLGTPKKEINRVLYSLAKKGKLQKE 188
> 7303451
Length=816
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 26/90 (28%), Positives = 40/90 (44%), Gaps = 14/90 (15%)
Query 67 LASEEKAFTER--ELIDRLGWPDKRVREVCASLERLMLIQKEQLSAASAQALSNASADGD 124
L +E +A +R EL LG R++ LE LM + K Q +A+ +++ G
Sbjct 472 LINELRALRQRKGELEGHLGALQDSRRQLMEQLEGLMRMLKNQQTASPRSTPNSSPRSGK 531
Query 125 TTSSVGGTTDIGRGSMPGGAALLGSGASSA 154
+ MPGGA +LG+ SA
Sbjct 532 SPP------------MPGGAGILGTSPMSA 549
> Hs7669475
Length=1181
Score = 30.0 bits (66), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query 54 LFCDDEQIVVVDFLAS--EEKAFTERELIDRLGWPDKRVREVCASLERLMLIQKE 106
++ D EQ ++ FL E KA T +L +LG P K + V SL + +QKE
Sbjct 135 IYQDQEQ-RILKFLEELGEGKATTAHDLSGKLGTPKKEINRVLYSLAKKGKLQKE 188
> Hs7669473
Length=1200
Score = 30.0 bits (66), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query 54 LFCDDEQIVVVDFLAS--EEKAFTERELIDRLGWPDKRVREVCASLERLMLIQKE 106
++ D EQ ++ FL E KA T +L +LG P K + V SL + +QKE
Sbjct 135 IYQDQEQ-RILKFLEELGEGKATTAHDLSGKLGTPKKEINRVLYSLAKKGKLQKE 188
> Hs4506807
Length=1836
Score = 28.9 bits (63), Expect = 6.5, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 26/58 (44%), Gaps = 3/58 (5%)
Query 97 LERLMLIQKEQLSAASAQALSNASADGDTT--SSVGGTTDIGRGSMPGGAALLGSGAS 152
LE+ Q+E A +AQAL ADGD G+ D +G G GSG S
Sbjct 472 LEKFKKHQEELEKAKAAQALEGGEADGDPAHGKDCNGSLDTSQGE-KGAPRQSGSGDS 528
Lambda K H
0.316 0.130 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3515233148
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40