bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2052_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
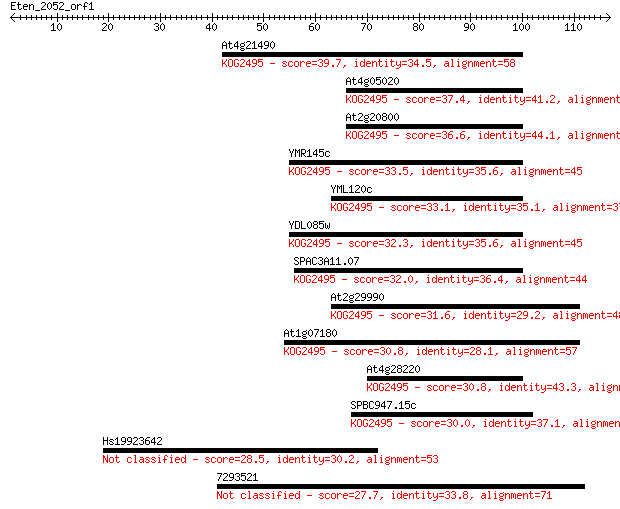
At4g21490 39.7 0.001
At4g05020 37.4 0.007
At2g20800 36.6 0.012
YMR145c 33.5 0.10
YML120c 33.1 0.14
YDL085w 32.3 0.20
SPAC3A11.07 32.0 0.26
At2g29990 31.6 0.41
At1g07180 30.8 0.58
At4g28220 30.8 0.60
SPBC947.15c 30.0 1.0
Hs19923642 28.5 2.8
7293521 27.7 5.7
> At4g21490
Length=478
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 34/61 (55%), Gaps = 3/61 (4%)
Query 42 LLHYLGFIKPAAAADPQQQQ---QQQQRRKRVVVCGGGWAAASFISKLDYFKFEVLLLLP 98
L H G + + A+P + + R+++VV+ G GWA ASF+ L+ +EV ++ P
Sbjct 15 LFHSGGGLIVYSEANPSYSNNGVETKTRKRKVVLLGTGWAGASFLKTLNNSSYEVQVISP 74
Query 99 R 99
R
Sbjct 75 R 75
> At4g05020
Length=583
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 24/34 (70%), Gaps = 0/34 (0%)
Query 66 RRKRVVVCGGGWAAASFISKLDYFKFEVLLLLPR 99
++K+VV+ G GWA SF+ L+ ++EV ++ PR
Sbjct 57 KKKKVVLLGTGWAGTSFLKNLNNSQYEVQIISPR 90
> At2g20800
Length=582
Score = 36.6 bits (83), Expect = 0.012, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 24/34 (70%), Gaps = 0/34 (0%)
Query 66 RRKRVVVCGGGWAAASFISKLDYFKFEVLLLLPR 99
R+K+VVV G GW+ SF+S L+ ++V ++ PR
Sbjct 62 RKKKVVVLGSGWSGYSFLSYLNNPNYDVQVVSPR 95
> YMR145c
Length=560
Score = 33.5 bits (75), Expect = 0.10, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 26/51 (50%), Gaps = 6/51 (11%)
Query 55 ADPQQQQQQQ------QRRKRVVVCGGGWAAASFISKLDYFKFEVLLLLPR 99
A+P Q Q +RK +V+ G GW + S + LD + V+++ PR
Sbjct 94 ANPSTQVPQSDTFPNGSKRKTLVILGSGWGSVSLLKNLDTTLYNVVVVSPR 144
> YML120c
Length=513
Score = 33.1 bits (74), Expect = 0.14, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 63 QQQRRKRVVVCGGGWAAASFISKLDYFKFEVLLLLPR 99
Q + V++ G GW A SF+ +D K+ V ++ PR
Sbjct 49 QHSDKPNVLILGSGWGAISFLKHIDTKKYNVSIISPR 85
> YDL085w
Length=545
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 6/51 (11%)
Query 55 ADPQQQQQQQQ------RRKRVVVCGGGWAAASFISKLDYFKFEVLLLLPR 99
++P +Q Q ++K +V+ G GW A S + KLD + V ++ PR
Sbjct 79 SNPPKQVPQSTAFANGLKKKELVILGTGWGAISLLKKLDTSLYNVTVVSPR 129
> SPAC3A11.07
Length=551
Score = 32.0 bits (71), Expect = 0.26, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 1/44 (2%)
Query 56 DPQQQQQQQQRRKRVVVCGGGWAAASFISKLDYFKFEVLLLLPR 99
DP Q + K +VV G GW A S + +D F V+++ PR
Sbjct 81 DPHQPLPDPSK-KTLVVLGAGWGATSILRTIDTSLFNVIVVSPR 123
> At2g29990
Length=508
Score = 31.6 bits (70), Expect = 0.41, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 63 QQQRRKRVVVCGGGWAAASFISKLDYFKFEVLLLLPRQQQQTHPAAAA 110
++ + RVVV G GWA + +D ++V+ + PR P A+
Sbjct 67 REGEKPRVVVLGSGWAGCRLMKGIDTNLYDVVCVSPRNHMVFTPLLAS 114
> At1g07180
Length=510
Score = 30.8 bits (68), Expect = 0.58, Method: Composition-based stats.
Identities = 16/61 (26%), Positives = 29/61 (47%), Gaps = 4/61 (6%)
Query 54 AADPQQQQ----QQQQRRKRVVVCGGGWAAASFISKLDYFKFEVLLLLPRQQQQTHPAAA 109
A +PQ+ ++ + RV+V G GWA + +D ++V+ + PR P A
Sbjct 56 ALEPQRYDGLAPTKEGEKPRVLVLGSGWAGCRVLKGIDTSIYDVVCVSPRNHMVFTPLLA 115
Query 110 A 110
+
Sbjct 116 S 116
> At4g28220
Length=559
Score = 30.8 bits (68), Expect = 0.60, Method: Composition-based stats.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 70 VVVCGGGWAAASFISKLDYFKFEVLLLLPR 99
VVV G GWA SF+ LD ++V ++ P+
Sbjct 52 VVVLGTGWAGISFLKDLDITSYDVQVVSPQ 81
> SPBC947.15c
Length=551
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 67 RKRVVVCGGGWAAASFISKLDYFKFEVLLLLPRQQ 101
+K +VV G GW A + I LD + + L+ PR
Sbjct 90 KKNIVVLGSGWGAVAAIKNLDPSLYNITLVSPRDH 124
> Hs19923642
Length=1004
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 16/53 (30%), Positives = 30/53 (56%), Gaps = 1/53 (1%)
Query 19 ASPPRPPGVCTLQLLQLRLRLQSLLHYLGFIKPAAAADPQQQQQQQQRRKRVV 71
A PP VC L++ QL + L+++ H+ I A+P+ +++Q KR++
Sbjct 416 ACPPVSMDVCALRI-QLFIGLKAICHFKNHIILLTKAEPEAIPERRQSPKRLL 467
> 7293521
Length=1723
Score = 27.7 bits (60), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 34/74 (45%), Gaps = 3/74 (4%)
Query 41 SLLHYLGFIKPAAAADPQQQQQQQQRRKRVV---VCGGGWAAASFISKLDYFKFEVLLLL 97
S++ + G K AA+ PQ QQQ ++R RVV + AS S+ D E +
Sbjct 116 SVVAHSGRTKSPAASQPQLQQQMKKRPTRVVPGTTTPRRVSDASMASESDSDSDEPVRRP 175
Query 98 PRQQQQTHPAAAAA 111
RQ + P A A
Sbjct 176 KRQSAKDKPQAGKA 189
Lambda K H
0.323 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174970866
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40