bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2048_orf2
Length=180
Score E
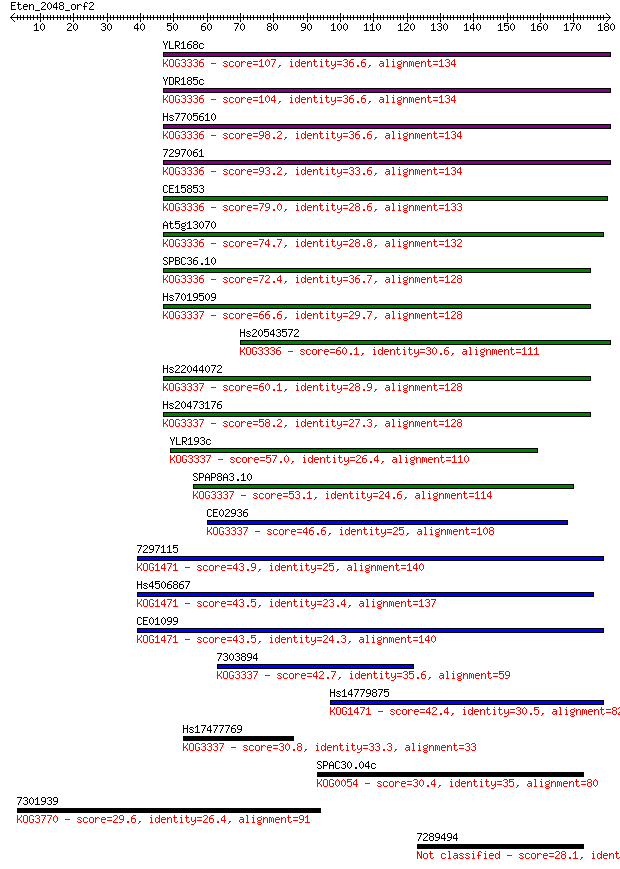
Sequences producing significant alignments: (Bits) Value
YLR168c 107 1e-23
YDR185c 104 9e-23
Hs7705610 98.2 8e-21
7297061 93.2 2e-19
CE15853 79.0 4e-15
At5g13070 74.7 9e-14
SPBC36.10 72.4 4e-13
Hs7019509 66.6 2e-11
Hs20543572 60.1 2e-09
Hs22044072 60.1 3e-09
Hs20473176 58.2 9e-09
YLR193c 57.0 2e-08
SPAP8A3.10 53.1 3e-07
CE02936 46.6 3e-05
7297115 43.9 2e-04
Hs4506867 43.5 2e-04
CE01099 43.5 2e-04
7303894 42.7 4e-04
Hs14779875 42.4 6e-04
Hs17477769 30.8 1.5
SPAC30.04c 30.4 1.7
7301939 29.6 3.0
7289494 28.1 9.8
> YLR168c
Length=230
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 49/135 (36%), Positives = 76/135 (56%), Gaps = 1/135 (0%)
Query 47 MRLFQKEHVFHNDWDTITSAFWVKYPNELQPHVLRVDTLDLLLDTEQQQFSCRRLLSLKY 106
M+LFQ + F+ WD +T+A W KYPNE+ HV+ VD L L + + RL+++K
Sbjct 1 MKLFQNSYDFNYPWDQVTAANWKKYPNEISTHVIAVDVLRRELKDQGKVLVTERLITVKQ 60
Query 107 QCPKWVQKFFGASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPEN-P 165
PKW+ G + + + E + L K LT++SCN T + +V ET Y PHP++
Sbjct 61 GVPKWIMMMLGGTNMSHVREVSVVDLNKKSLTMRSCNLTMCNLLKVYETVTYSPHPDDSA 120
Query 166 QQTLYRQKATYRVSG 180
+TL++Q+A G
Sbjct 121 NKTLFQQEAQITAYG 135
> YDR185c
Length=179
Score = 104 bits (260), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 49/135 (36%), Positives = 77/135 (57%), Gaps = 2/135 (1%)
Query 47 MRLFQKEHVFHNDWDTITSAFWVKYPNELQPHVLRVDTLDLLLDTEQQQFSCRRLLSLKY 106
M+ FQK + F W+ +T+A W+KYPN++ HV+ VD L L RL++++
Sbjct 1 MKSFQKSYEFDYPWEKVTTANWMKYPNKISTHVIAVDVLRRELKEHGDVLLTERLITIRQ 60
Query 107 QCPKWVQKFFGASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPENPQ 166
P W+ G + + Y E +T +D+ LT++SCN TF + ET Y+PHP+NP
Sbjct 61 NTPHWMSILVGNTNLAYVREVSTVDRRDRSLTMRSCNMTFPHILKCYETVRYVPHPKNPS 120
Query 167 Q-TLYRQKATYRVSG 180
TL++Q A + +SG
Sbjct 121 NVTLFKQDAKF-LSG 134
> Hs7705610
Length=211
Score = 98.2 bits (243), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 49/135 (36%), Positives = 76/135 (56%), Gaps = 2/135 (1%)
Query 47 MRLFQKEHVFHNDWDTITSAFWVKYPNELQPHVLRVDTLDLLLDTEQQQFSCRRLLSLKY 106
M+++ EHVF + W+T+T+A KYPN + P V+ VD LD +D + S RLLS ++
Sbjct 1 MKIWTSEHVFDHPWETVTTAAMQKYPNPMNPSVVGVDVLDRHIDPSGKLHS-HRLLSTEW 59
Query 107 QCPKWVQKFFGASPV-GYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPENP 165
P V+ GA+ Y E + +K + LKS N +F + V+E Y PHP++P
Sbjct 60 GLPSIVKSLIGAARTKTYVQEHSVVDPVEKTMELKSTNISFTNMVSVDERLIYKPHPQDP 119
Query 166 QQTLYRQKATYRVSG 180
++T+ Q+A V G
Sbjct 120 EKTVLTQEAIITVKG 134
> 7297061
Length=215
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 75/135 (55%), Gaps = 4/135 (2%)
Query 47 MRLFQKEHVFHNDWDTITSAFWVKYPNELQPHVLRVDTLDL-LLDTEQQQFSCRRLLSLK 105
M+++ EH+F++ W+T+T A W KYPN + P ++ D ++ ++D RL+ K
Sbjct 1 MKIWTSEHIFNHPWETVTQAAWRKYPNPMTPSIIGTDVVERRVVD---GVLHTHRLVQSK 57
Query 106 YQCPKWVQKFFGASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPENP 165
+ PKW G + +A E++T + K++ LK+ N TF V+E Y PHP +
Sbjct 58 WYFPKWTHALIGTAKTCFASERSTVDPERKQMVLKTNNLTFCRNISVDEVLYYEPHPSDA 117
Query 166 QQTLYRQKATYRVSG 180
+TL +Q+AT V G
Sbjct 118 SKTLLKQEATVTVFG 132
> CE15853
Length=209
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 38/133 (28%), Positives = 65/133 (48%), Gaps = 2/133 (1%)
Query 47 MRLFQKEHVFHNDWDTITSAFWVKYPNELQPHVLRVDTLDLLLDTEQQQFSCRRLLSLKY 106
MR++ EH+F ++W+T+ A + KYPN L + +D + L E + R++ +
Sbjct 1 MRIWSSEHIFDHEWETVAQAAFRKYPNPLNRSITGIDVVKQTL--EAGKILTERIIQSHF 58
Query 107 QCPKWVQKFFGASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPENPQ 166
P W K G S Y+ E K+ +L + N +SF RV+E Y P E+P
Sbjct 59 SIPSWATKLTGFSGTQYSHEYTVIDPTRKEFSLTTRNLNGSSFLRVDEKLTYTPAHEDPN 118
Query 167 QTLYRQKATYRVS 179
+T+ +Q ++
Sbjct 119 KTILKQDVIVTIT 131
> At5g13070
Length=183
Score = 74.7 bits (182), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 38/137 (27%), Positives = 71/137 (51%), Gaps = 6/137 (4%)
Query 47 MRLFQKEHVFHNDWDTITSAFWVKYPN----ELQPHVLRVDTLDLLLDTEQQQFSCRRLL 102
++ +++EHV+ + W+ +++A W K+ + + H+L VDTL+ LDTE + R L
Sbjct 2 VKAYRQEHVYKHPWERVSAASWRKFADPENKRILSHILEVDTLNRKLDTETGKLHTTRAL 61
Query 103 SLKYQCPKWVQKFFGASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHP 162
++ P ++ + G + + E K + + L + N + F VEE Y PHP
Sbjct 62 TIHAPGPWFLHRIIGQD-ICHCVESTVVDGKSRSMQLTTKNISLKKFIEVEERIRYDPHP 120
Query 163 ENPQQ-TLYRQKATYRV 178
+NP T+ Q+ + R+
Sbjct 121 DNPSAWTVCSQETSIRI 137
> SPBC36.10
Length=184
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 47/129 (36%), Positives = 73/129 (56%), Gaps = 4/129 (3%)
Query 47 MRLFQKEHVFHNDWDTITSAFWVKYPNELQPHVLRVDTLDL-LLDTEQQQFSCRRLLSLK 105
M++F+ H+F ++ +++A W KYPNE HV+ VDTLD +LD RL++
Sbjct 1 MKIFESCHLFQYPFEQVSAAHWQKYPNEHATHVIAVDTLDRKVLDN--GVLYTERLITCH 58
Query 106 YQCPKWVQKFFGASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPENP 165
P+W+ K + Y E + LK + LTL + N TF+ RV+ET Y PHPE
Sbjct 59 QALPRWILKLIDGAQDCYIRETSYVDLKARTLTLLTSNLTFSDRLRVDETVTYSPHPE-L 117
Query 166 QQTLYRQKA 174
+ T+++Q+A
Sbjct 118 EATVFQQEA 126
> Hs7019509
Length=219
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 38/130 (29%), Positives = 65/130 (50%), Gaps = 3/130 (2%)
Query 47 MRLFQKEHVFHNDWDTITSAFWVKYPNELQPHVLRVDTLDLLLDTEQQQFSCRRLLSLKY 106
++ F + V + WD + +AFW +YPN HVL D + + +Q+ S RRLL+
Sbjct 2 VKYFLGQSVLRSSWDQVFAAFWQRYPNPYSKHVLTEDIVHREVTPDQKLLS-RRLLTKTN 60
Query 107 QCPKWVQKFFGASPVG--YAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPEN 164
+ P+W ++ F A+ Y E + +++ +T + N A VEE C Y + +N
Sbjct 61 RMPRWAERLFPANVAHSVYVLEDSIVDPQNQTMTTFTWNINHARLMVVEERCVYCVNSDN 120
Query 165 PQQTLYRQKA 174
T R++A
Sbjct 121 SGWTEIRREA 130
> Hs20543572
Length=143
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 55/112 (49%), Gaps = 2/112 (1%)
Query 70 KYPNELQPHVLRVDTLDLLLDTEQQQFSCRRLLSLKYQCPKWVQKFFGASPVG-YAFEQA 128
+ P +P V+ VD LD +D + S RLLS ++ P V+ GA+ + Y E +
Sbjct 11 EIPKPYEPTVVGVDVLDRHVDPSGKLHS-HRLLSTEWGLPSIVKSLIGAARMKTYVQEHS 69
Query 129 TCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPENPQQTLYRQKATYRVSG 180
+K + LKS N +F +E Y P P +P++T+ Q+A + G
Sbjct 70 VVDPVEKTMKLKSTNISFTDMVSADERLIYKPRPRDPEKTILTQEAIITMKG 121
> Hs22044072
Length=219
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 64/130 (49%), Gaps = 3/130 (2%)
Query 47 MRLFQKEHVFHNDWDTITSAFWVKYPNELQPHVLRVDTLDLLLDTEQQQFSCRRLLSLKY 106
++ F + V + WD + +AFW +YPN HVL D + + +Q+ S RLL+
Sbjct 2 VKYFLGQSVQRSSWDQVFAAFWQRYPNPYSKHVLTEDVVHREVTPDQKLLSG-RLLTKTN 60
Query 107 QCPKWVQKFFGASP--VGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPEN 164
+ P W ++ F A+ Y E + +++ +T + N A VEE C Y + +N
Sbjct 61 RTPCWAERLFPANVDHSVYILEDSIVDPQNQTMTTFTWNINHARLMVVEERCVYCVNSDN 120
Query 165 PQQTLYRQKA 174
+T R++A
Sbjct 121 SGRTEIRREA 130
> Hs20473176
Length=250
Score = 58.2 bits (139), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 35/130 (26%), Positives = 64/130 (49%), Gaps = 3/130 (2%)
Query 47 MRLFQKEHVFHNDWDTITSAFWVKYPNELQPHVLRVDTLDLLLDTEQQQFSCRRLLSLKY 106
++ F + V + WD + +AFW +YPN HVL DT+ + +Q+ S ++LL+
Sbjct 2 VKYFLGQGVLQSSWDQVFTAFWHQYPNPYSKHVLTEDTVHREVTADQKLLS-QQLLTKTN 60
Query 107 QCPKWVQKFFGASPVG--YAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPEN 164
+ W ++ F A+ + Y E + +++ +T + N A VE+ C Y + N
Sbjct 61 RMSHWAKQLFPANVLHSVYILEDSIVDPQNQTMTTFTWNINHARLMVVEKQCVYCMNSNN 120
Query 165 PQQTLYRQKA 174
T R++A
Sbjct 121 SGWTEIRREA 130
> YLR193c
Length=175
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/110 (26%), Positives = 51/110 (46%), Gaps = 1/110 (0%)
Query 49 LFQKEHVFHNDWDTITSAFWVKYPNELQPHVLRVDTLDLLLDTEQQQFSCRRLLSLKYQC 108
L + H+F D+ +++ AF+ +YPN PHVL +DT+ +D ++ RLL +
Sbjct 4 LHKSTHIFPTDFASVSRAFFNRYPNPYSPHVLSIDTISRNVD-QEGNLRTTRLLKKSGKL 62
Query 109 PKWVQKFFGASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEY 158
P WV+ F + E + + + + + N +VEE Y
Sbjct 63 PTWVKPFLRGITETWIIEVSVVNPANSTMKTYTRNLDHTGIMKVEEYTTY 112
> SPAP8A3.10
Length=171
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/114 (24%), Positives = 54/114 (47%), Gaps = 2/114 (1%)
Query 56 FHNDWDTITSAFWVKYPNELQPHVLRVDTLDLLLDTEQQQFSCRRLLSLKYQCPKWVQKF 115
+ W+T++SA+ +YPN HV+ D L+ +D E + ++ RLL + + P+W
Sbjct 11 LNASWNTVSSAWLTRYPNPYSLHVVSADVLERYVDDEGRLYT-ERLLVKQGRLPRWASDL 69
Query 116 FGASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPENPQQTL 169
+ Y E++ ++L ++ N RV E ++ EN +T+
Sbjct 70 LNVNK-SYILERSVIDPSKQELKSETFNLDHVKILRVIEYSRFIQSSENCSKTI 122
> CE02936
Length=210
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 27/111 (24%), Positives = 50/111 (45%), Gaps = 4/111 (3%)
Query 60 WDTITSAFWVKYPNELQPHVLRVDTLDLLLDTE---QQQFSCRRLLSLKYQCPKWVQKFF 116
+D + SAFW +YPN H++ D L+ + ++ ++ S+ + P+W+ +
Sbjct 15 FDEVASAFWDRYPNSHAKHIISEDVLERQITDNTIVTKKLIVKQGSSILKRVPRWISRMT 74
Query 117 GASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPENPQQ 167
V E++ KKL + N + S F++ E C Y +N Q
Sbjct 75 DIQVVP-VIEESVYDKVSKKLVTYTRNVSHISLFQLHERCIYKSSEDNQQH 124
> 7297115
Length=321
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 68/142 (47%), Gaps = 10/142 (7%)
Query 39 LLQACEAPMRLFQKEHVFHNDWDTITSAFWVKYPNELQ-PHVLRVDTL-DLLLDTEQQQF 96
++Q ++P+R V+ ++ + A+ ++P Q P VL + + D L+ ++
Sbjct 1 MVQKFQSPVR------VYKYPFELVMKAYERRFPTCPQMPIVLDCEVIKDESLEDGAKRN 54
Query 97 SCRRLLSLKYQCPKWVQKFFGASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETC 156
+ RR L P +K G V Y + L ++ L++++ N +F+S + E C
Sbjct 55 TSRRC-KLAVDAPYIFKKLIGVDHV-YFLQHNFLDLANRTLSIEAVNESFSSRIEIFERC 112
Query 157 EYLPHPENPQQTLYRQKATYRV 178
Y HP+N + T + Q AT +
Sbjct 113 RYYAHPDNSEWTCFDQSATLDI 134
> Hs4506867
Length=715
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 32/138 (23%), Positives = 65/138 (47%), Gaps = 8/138 (5%)
Query 39 LLQACEAPMRLFQKEHVFHNDWDTITSAFWVKYPN-ELQPHVLRVDTLDLLLDTEQQQFS 97
++Q ++P+R V+ ++ I +A+ ++P L P + DT+ +
Sbjct 1 MVQKYQSPVR------VYKYPFELIMAAYERRFPTCPLIPMFVGSDTVSEFKSEDGAIHV 54
Query 98 CRRLLSLKYQCPKWVQKFFGASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCE 157
R L P+ ++K G V Y ++ + + +++ L +++ N TF++ + E C
Sbjct 55 IERRCKLDVDAPRLLKKIAGVDYV-YFVQKNSLNSRERTLHIEAYNETFSNRVIINEHCC 113
Query 158 YLPHPENPQQTLYRQKAT 175
Y HPEN T + Q A+
Sbjct 114 YTVHPENEDWTCFEQSAS 131
> CE01099
Length=743
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/150 (22%), Positives = 68/150 (45%), Gaps = 26/150 (17%)
Query 39 LLQACEAPMRLFQKEHVFHNDWDTITSAFWVKYP----------NELQPHVLRVDTLDLL 88
++Q +P+R+++ H F + + +A+ +++P +E+ VD + +
Sbjct 1 MVQTYRSPVRIYK--HPF----EIVMAAYEMRFPTCPQIPIFVGSEVTYEYKSVDGAEWV 54
Query 89 LDTEQQQFSCRRLLSLKYQCPKWVQKFFGASPVGYAFEQATCSLKDKKLTLKSCNFTFAS 148
+D R L + P V+K G V Y ++ + + + L +++ N +F+S
Sbjct 55 ID---------RKCQLNVEAPYLVKKIAGVDYV-YFSQKNSLDRRKRTLDIEATNISFSS 104
Query 149 FFRVEETCEYLPHPENPQQTLYRQKATYRV 178
V+E C Y H EN T + Q A+ V
Sbjct 105 RINVKENCTYYVHAENENWTCFEQSASLDV 134
> 7303894
Length=171
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 29/59 (49%), Gaps = 2/59 (3%)
Query 63 ITSAFWVKYPNELQPHVLRVDTLDLLLDTEQQQFSCRRLLSLKYQCPKWVQKFFGASPV 121
+ A+W +YPN HVL DT+ + + RRLLS PKW +F+ PV
Sbjct 3 VVVAYWNRYPNPSSTHVLTEDTIQ--REVRDGKLFSRRLLSKTNPVPKWGARFYNNVPV 59
> Hs14779875
Length=696
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 41/82 (50%), Gaps = 5/82 (6%)
Query 97 SCRRLLSLKYQCPKWVQKFFGASPVGYAFEQATCSLKDKKLTLKSCNFTFASFFRVEETC 156
SCR L+ P+ ++K G V + + + K++ L +++ N TFA+ V E C
Sbjct 58 SCR----LRVDAPRLLRKIAGVEHVVFV-QTNILNWKERTLLIEAHNETFANRVVVNEHC 112
Query 157 EYLPHPENPQQTLYRQKATYRV 178
Y HPEN T + Q A+ +
Sbjct 113 SYTVHPENEDWTCFEQSASLDI 134
> Hs17477769
Length=168
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 53 EHVFHNDWDTITSAFWVKYPNELQPHVLRVDTL 85
+ V + WD + +AFW +YPN +L D +
Sbjct 8 QGVLRSSWDQVFAAFWQRYPNPCGKRILTEDIV 40
> SPAC30.04c
Length=1469
Score = 30.4 bits (67), Expect = 1.7, Method: Composition-based stats.
Identities = 28/106 (26%), Positives = 36/106 (33%), Gaps = 26/106 (24%)
Query 93 QQQFSCR-----RLLSLKYQCPKWV---------------------QKFFGASPVGYAFE 126
+Q FS R RL S Y +WV QK VG++
Sbjct 1096 KQMFSERLDNLVRLQSTSYNLNRWVAVRTDGISGLVGAIAGLIALLQKDLSPGVVGFSLN 1155
Query 127 QATCSLKDKKLTLKSCNFTFASFFRVEETCEYLPHPENPQQTLYRQ 172
QA L ++SCN A E EY P+ P T+ Q
Sbjct 1156 QAVIFSSSVLLFVRSCNSLQAEMNSYERVLEYSKLPQEPAPTIAGQ 1201
> 7301939
Length=638
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 24/97 (24%), Positives = 40/97 (41%), Gaps = 16/97 (16%)
Query 3 QGLESAALRPRCCRTSFCKFPRSLTHASFAATTSPQLLQACEAPMRLFQK--EHVFHNDW 60
+G +A P CCR S + S A F + + C+ P RL EH+ N
Sbjct 201 EGSNAACDEPMCCRNSLPEGSDSSAAAGFWSD-----YRDCDCPKRLILSAFEHIKENH- 254
Query 61 DTITSAFWVKYPNELQPHVL----RVDTLDLLLDTEQ 93
W+ + ++ PH + R LD+L + ++
Sbjct 255 ----KIEWIYHTGDVPPHNVWSTTRQGNLDMLSEIDE 287
> 7289494
Length=674
Score = 28.1 bits (61), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 26/54 (48%), Gaps = 4/54 (7%)
Query 123 YAFEQATCSLKDKKLTLKSCNFTFASFFRVEETCE----YLPHPENPQQTLYRQ 172
Y + D+ L + + NF ++ R++ E Y HP++ +Q LYR+
Sbjct 357 YCISFGMSEMNDQTLRISTPNFAPSALMRLQHNAEALQEYKKHPDSLRQKLYRE 410
Lambda K H
0.325 0.134 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2806646388
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40