bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2042_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
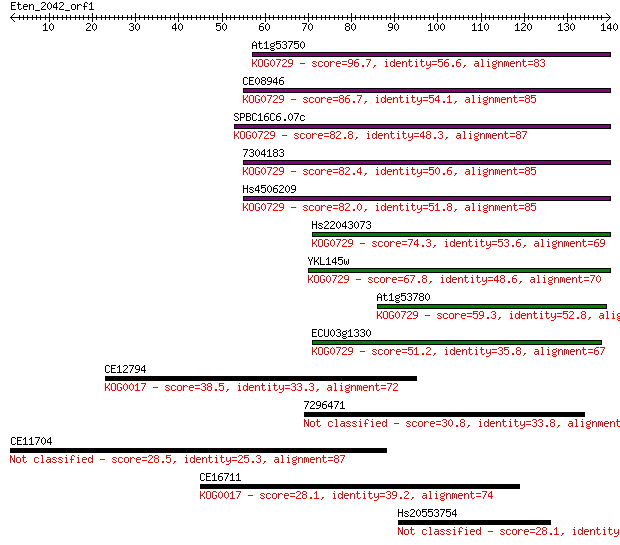
At1g53750 96.7 1e-20
CE08946 86.7 1e-17
SPBC16C6.07c 82.8 2e-16
7304183 82.4 2e-16
Hs4506209 82.0 3e-16
Hs22043073 74.3 7e-14
YKL145w 67.8 6e-12
At1g53780 59.3 2e-09
ECU03g1330 51.2 7e-07
CE12794 38.5 0.004
7296471 30.8 0.72
CE11704 28.5 4.4
CE16711 28.1 4.9
Hs20553754 28.1 5.6
> At1g53750
Length=426
Score = 96.7 bits (239), Expect = 1e-20, Method: Composition-based stats.
Identities = 47/84 (55%), Positives = 61/84 (72%), Gaps = 4/84 (4%)
Query 57 DESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDT 116
DE +KN P PLD DI +LK+YGL PY+ KK+E +IK + I+ LCG++ESDT
Sbjct 7 DEIRDEKN---PRPLDEDDIALLKTYGLGPYSAPIKKVEKEIKDLAKKINDLCGIKESDT 63
Query 117 GLSLPSQWDLAADKRLMQ-EQPLQ 139
GL+ PSQWDL +DK++MQ EQPLQ
Sbjct 64 GLAPPSQWDLVSDKQMMQEEQPLQ 87
> CE08946
Length=435
Score = 86.7 bits (213), Expect = 1e-17, Method: Composition-based stats.
Identities = 46/86 (53%), Positives = 60/86 (69%), Gaps = 4/86 (4%)
Query 55 KGDESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRES 114
K D E++KN A LD DI +LK YG PYA K L+ADI+ L+ +++L GV+ES
Sbjct 14 KDDTKEEEKNFQA---LDEGDIAVLKRYGQGPYAEQLKTLDADIENCLKKVNELSGVKES 70
Query 115 DTGLSLPSQWDLAADKRLM-QEQPLQ 139
DTGL+ P+ WD+AADK+ M QEQPLQ
Sbjct 71 DTGLAPPALWDIAADKQAMQQEQPLQ 96
> SPBC16C6.07c
Length=438
Score = 82.8 bits (203), Expect = 2e-16, Method: Composition-based stats.
Identities = 42/88 (47%), Positives = 57/88 (64%), Gaps = 4/88 (4%)
Query 53 RPKGDESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVR 112
+P E E DKN PPPLD DI++LKSY PYA ++ D + L+ I+ G++
Sbjct 12 KPVDTEEENDKN---PPPLDEGDIELLKSYATGPYARELAAIKEDTEAVLKRINDTVGIK 68
Query 113 ESDTGLSLPSQWDLAADK-RLMQEQPLQ 139
ESDTGL+ S WD+AAD+ R+ +EQPLQ
Sbjct 69 ESDTGLAPISFWDVAADRQRMSEEQPLQ 96
> 7304183
Length=433
Score = 82.4 bits (202), Expect = 2e-16, Method: Composition-based stats.
Identities = 43/86 (50%), Positives = 62/86 (72%), Gaps = 5/86 (5%)
Query 55 KGDESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRES 114
K DE E DK + LD DI++LK+YG S Y S K +E DI++ ++ +++L G++ES
Sbjct 13 KHDEKE-DKEIKS---LDEGDIELLKTYGQSQYHKSIKSIEEDIQKAVKQVNELTGIKES 68
Query 115 DTGLSLPSQWDLAADKRLMQ-EQPLQ 139
DTGL+ P+ WDLAADK+++Q EQPLQ
Sbjct 69 DTGLAPPALWDLAADKQILQNEQPLQ 94
> Hs4506209
Length=433
Score = 82.0 bits (201), Expect = 3e-16, Method: Composition-based stats.
Identities = 44/86 (51%), Positives = 59/86 (68%), Gaps = 3/86 (3%)
Query 55 KGDESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRES 114
K E EKD LD DI +LK+YG S Y+ K++E DI+Q L+ I++L G++ES
Sbjct 11 KTKEDEKDDKPIRA--LDEGDIALLKTYGQSTYSRQIKQVEDDIQQLLKKINELTGIKES 68
Query 115 DTGLSLPSQWDLAADKRLMQ-EQPLQ 139
DTGL+ P+ WDLAADK+ +Q EQPLQ
Sbjct 69 DTGLAPPALWDLAADKQTLQSEQPLQ 94
> Hs22043073
Length=318
Score = 74.3 bits (181), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 37/70 (52%), Positives = 53/70 (75%), Gaps = 1/70 (1%)
Query 71 LDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWDLAADK 130
LD ADI +LK+YG S Y K++E ++Q L+ I++L G++ESDTGL+ P+ WDLAADK
Sbjct 25 LDEADIALLKTYGHSTYFRQIKQVEDGVQQLLKKINELTGIKESDTGLAPPALWDLAADK 84
Query 131 RLMQ-EQPLQ 139
+ +Q +QPLQ
Sbjct 85 QTLQSKQPLQ 94
> YKL145w
Length=467
Score = 67.8 bits (164), Expect = 6e-12, Method: Composition-based stats.
Identities = 34/71 (47%), Positives = 44/71 (61%), Gaps = 1/71 (1%)
Query 70 PLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWDLAAD 129
PL DI +LKSYG +PYA K+ E D+K I + GV+ESDTGL+ WD+ D
Sbjct 28 PLTEGDIQVLKSYGAAPYAAKLKQTENDLKDIEARIKEKAGVKESDTGLAPSHLWDIMGD 87
Query 130 K-RLMQEQPLQ 139
+ RL +E PLQ
Sbjct 88 RQRLGEEHPLQ 98
> At1g53780
Length=464
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 28/54 (51%), Positives = 40/54 (74%), Gaps = 2/54 (3%)
Query 86 PYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWDLAADKRLMQ-EQPL 138
PY+ KK+E +I + + I L G++ESDTGL+ P+QWDL +DK++MQ EQPL
Sbjct 71 PYSARIKKVEKEINELAEKICNL-GIKESDTGLAPPNQWDLVSDKQMMQEEQPL 123
> ECU03g1330
Length=415
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 39/67 (58%), Gaps = 0/67 (0%)
Query 71 LDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWDLAADK 130
++ D+ + SYG + YA + EA+I++ + IS G RE +TGL+ P WDL D+
Sbjct 10 IEEKDVGFILSYGRNYYADGIVEKEAEIRRLFEQISLKLGTREVETGLAPPMCWDLLGDR 69
Query 131 RLMQEQP 137
+Q +P
Sbjct 70 NRIQSEP 76
> CE12794
Length=1455
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 35/72 (48%), Gaps = 1/72 (1%)
Query 23 ALLRVCCPRWLHYLLSLLGAAREMANPSSSRPKGDESEKDKNTAAPPPLDAADIDILKSY 82
A +R ++H + L AA E N DESE ++ + PPP AD +I SY
Sbjct 372 ATVRQHLASFMHDVTERLRAADE-GNACPQESVVDESEAEEERSGPPPTPQADTNIHSSY 430
Query 83 GLSPYAGSTKKL 94
G+ + +TK L
Sbjct 431 GVINFETNTKNL 442
> 7296471
Length=1455
Score = 30.8 bits (68), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 33/69 (47%), Gaps = 8/69 (11%)
Query 69 PPLDAADIDI----LKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQW 124
P L + D I SYGLS +T LE +LQ + + VR++ TGL +P+
Sbjct 249 PKLASGDFTIDQVMRSSYGLS----ATINLEGSSANKLQTLPTVTLVRDAQTGLEVPTYA 304
Query 125 DLAADKRLM 133
L + L+
Sbjct 305 QLNQNNSLL 313
> CE11704
Length=117
Score = 28.5 bits (62), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 37/87 (42%), Gaps = 0/87 (0%)
Query 1 TLVKELLLLLGDLCTYTWSCVAALLRVCCPRWLHYLLSLLGAAREMANPSSSRPKGDESE 60
++ + L L + L + + + P+ H + EM+ S+ R + DE +
Sbjct 6 SINRGLGLGMAQLGAFARMGMMINVSAMGPQDGHANGGVGEGGIEMSRKSTWRSENDEIK 65
Query 61 KDKNTAAPPPLDAADIDILKSYGLSPY 87
K+TA P LD ID + G S Y
Sbjct 66 TRKSTAFEPDLDFDMIDFRQPSGSSQY 92
> CE16711
Length=2356
Score = 28.1 bits (61), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 39/80 (48%), Gaps = 10/80 (12%)
Query 45 EMANPSSSRPKGDESEKDKNTAAPPPLDAADIDILK---SY-GLSP--YAGSTKKLEADI 98
EM +PS+ + K EK K P A + ILK S+ + P Y S +KL +
Sbjct 1745 EMIDPSTKKLKKPLMEKAKK----PSATATVLHILKLPSSFESIVPFKYTNSLRKLMLIV 1800
Query 99 KQQLQNISKLCGVRESDTGL 118
+ L ISK VR SDTG+
Sbjct 1801 YRILTFISKTIPVRASDTGV 1820
> Hs20553754
Length=809
Score = 28.1 bits (61), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 17/35 (48%), Gaps = 0/35 (0%)
Query 91 TKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWD 125
T LE IK LQ I ++ E D+ P QWD
Sbjct 774 TSDLERSIKAALQRIKRVSADSEEDSDEQDPGQWD 808
Lambda K H
0.316 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40