bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2040_orf1
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
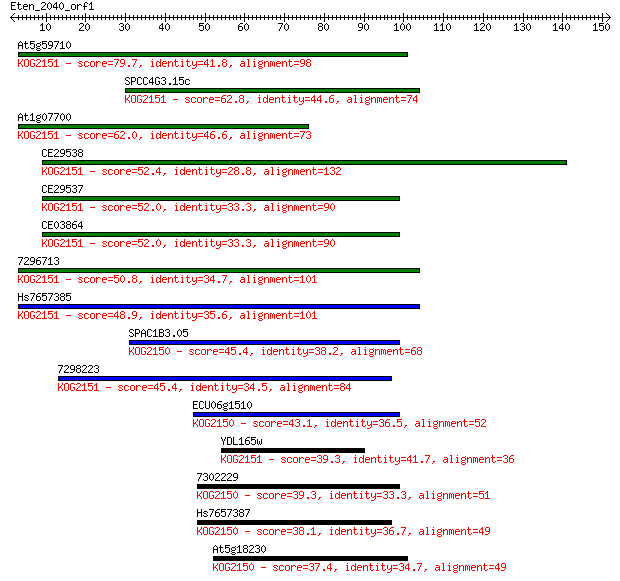
At5g59710 79.7 2e-15
SPCC4G3.15c 62.8 2e-10
At1g07700 62.0 4e-10
CE29538 52.4 3e-07
CE29537 52.0 4e-07
CE03864 52.0 4e-07
7296713 50.8 9e-07
Hs7657385 48.9 4e-06
SPAC1B3.05 45.4 4e-05
7298223 45.4 4e-05
ECU06g1510 43.1 2e-04
YDL165w 39.3 0.003
7302229 39.3 0.003
Hs7657387 38.1 0.007
At5g18230 37.4 0.010
> At5g59710
Length=616
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 41/98 (41%), Positives = 49/98 (50%), Gaps = 0/98 (0%)
Query 3 LNSTECLYGAFTSPISETPVTKTVEELLPQCYVAAAPTPFRTSHLEKLETRTLFYIFYTS 62
LNST LY F SP + P VE +P CY A P P + ++ LFY FY+
Sbjct 479 LNSTGNLYKTFASPWTNEPAKSEVEFTVPNCYYATEPPPLTRASFKRFSYELLFYTFYSM 538
Query 63 PRLLLQGYAAVELTRRQWTYHKEWRKWFRAAEAPGVSS 100
P+ Q YAA EL R W YHKE R WF P V +
Sbjct 539 PKDEAQLYAADELYERGWFYHKELRVWFFRVGEPLVRA 576
> SPCC4G3.15c
Length=176
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 41/74 (55%), Gaps = 3/74 (4%)
Query 30 LPQCYVAAAPTPFRTSHLEKLETRTLFYIFYTSPRLLLQGYAAVELTRRQWTYHKEWRKW 89
LP CY P P S + + TLFYIFYT PR ++Q AA ELT R W +HKE R W
Sbjct 65 LPACYKNVNPPP-AISKIFQFSDETLFYIFYTMPRDVMQEAAAQELTNRNWRFHKELRVW 123
Query 90 FRAAEAPGVSSLRK 103
PG+ L++
Sbjct 124 L--TPVPGMKPLQR 135
> At1g07700
Length=759
Score = 62.0 bits (149), Expect = 4e-10, Method: Composition-based stats.
Identities = 34/73 (46%), Positives = 39/73 (53%), Gaps = 0/73 (0%)
Query 3 LNSTECLYGAFTSPISETPVTKTVEELLPQCYVAAAPTPFRTSHLEKLETRTLFYIFYTS 62
LNSTE L+ F SP S P E +PQCY A P P KL TLFY+FY+
Sbjct 687 LNSTENLHKTFGSPWSNEPSKVDPEFSVPQCYYAKNPPPLHQGLFAKLLVETLFYVFYSM 746
Query 63 PRLLLQGYAAVEL 75
P+ Q YAA EL
Sbjct 747 PKDEAQLYAANEL 759
> CE29538
Length=317
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 38/138 (27%), Positives = 62/138 (44%), Gaps = 9/138 (6%)
Query 9 LYGAFTSPISETPV------TKTVEELLPQCYVAAAPTPFRTSHLEKLETRTLFYIFYTS 62
LY F P +++P+ K EE + ++ P R L K+ LFY+FY
Sbjct 97 LYMNFGGPWADSPIRAHELDVKVPEEYMTHNHIRDKLPPLR---LNKVSEDVLFYLFYNC 153
Query 63 PRLLLQGYAAVELTRRQWTYHKEWRKWFRAAEAPGVSSLRKRLPEGGDPLQQQQQQQQQD 122
P + Q AA EL R+W +HK + W ++ GV +G + Q Q ++
Sbjct 154 PNEIYQVAAACELYAREWRFHKSEQVWLTRSQYGGVKEQTGNYEKGHYNVFDQMQWRKIP 213
Query 123 QQQQQGYKEAELNQELQQ 140
++ + YKE E ++ Q
Sbjct 214 KELKLEYKELEDRPKMPQ 231
> CE29537
Length=367
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 45/96 (46%), Gaps = 9/96 (9%)
Query 9 LYGAFTSPISETPV------TKTVEELLPQCYVAAAPTPFRTSHLEKLETRTLFYIFYTS 62
LY F P +++P+ K EE + ++ P R L K+ LFY+FY
Sbjct 147 LYMNFGGPWADSPIRAHELDVKVPEEYMTHNHIRDKLPPLR---LNKVSEDVLFYLFYNC 203
Query 63 PRLLLQGYAAVELTRRQWTYHKEWRKWFRAAEAPGV 98
P + Q AA EL R+W +HK + W ++ GV
Sbjct 204 PNEIYQVAAACELYAREWRFHKSEQVWLTRSQYGGV 239
> CE03864
Length=444
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 45/96 (46%), Gaps = 9/96 (9%)
Query 9 LYGAFTSPISETPV------TKTVEELLPQCYVAAAPTPFRTSHLEKLETRTLFYIFYTS 62
LY F P +++P+ K EE + ++ P R L K+ LFY+FY
Sbjct 224 LYMNFGGPWADSPIRAHELDVKVPEEYMTHNHIRDKLPPLR---LNKVSEDVLFYLFYNC 280
Query 63 PRLLLQGYAAVELTRRQWTYHKEWRKWFRAAEAPGV 98
P + Q AA EL R+W +HK + W ++ GV
Sbjct 281 PNEIYQVAAACELYAREWRFHKSEQVWLTRSQYGGV 316
> 7296713
Length=579
Score = 50.8 bits (120), Expect = 9e-07, Method: Composition-based stats.
Identities = 35/104 (33%), Positives = 49/104 (47%), Gaps = 5/104 (4%)
Query 3 LNSTECLYGAFTSPISETPV-TKTVEELLPQCYVA--AAPTPFRTSHLEKLETRTLFYIF 59
LNS E L+ F P P + VE +P Y+ A L+KL+ LF++F
Sbjct 432 LNSQESLHTTFAGPFVAQPCRAQDVEFNVPPEYLINFAIRDKLTAPVLKKLQEDLLFFLF 491
Query 60 YTSPRLLLQGYAAVELTRRQWTYHKEWRKWFRAAEAPGVSSLRK 103
YT+ ++Q AA EL R+W YH E + W PG+ K
Sbjct 492 YTNIGDMMQLMAAAELHSREWRYHVEEKIWI--TRIPGIDQYEK 533
> Hs7657385
Length=540
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 36/104 (34%), Positives = 51/104 (49%), Gaps = 5/104 (4%)
Query 3 LNSTECLYGAFTSPISETPV-TKTVEELLPQCYVAAAPTPFRTS--HLEKLETRTLFYIF 59
LNS E LY F SP + +P + ++ +P Y+ + + L + LFY++
Sbjct 389 LNSPENLYPKFASPWASSPCRPQDIDFHVPSEYLTNIHIRDKLAAIKLGRYGEDLLFYLY 448
Query 60 YTSPRLLLQGYAAVELTRRQWTYHKEWRKWFRAAEAPGVSSLRK 103
Y + +LQ AAVEL R W YHKE R W APG+ K
Sbjct 449 YMNGGDVLQLLAAVELFNRDWRYHKEERVWI--TRAPGMEPTMK 490
> SPAC1B3.05
Length=630
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 39/81 (48%), Gaps = 13/81 (16%)
Query 31 PQCYVAAAPTP------------FRTSHL-EKLETRTLFYIFYTSPRLLLQGYAAVELTR 77
PQ Y+ P P F +S + E ++ TLFY+FY P Q A EL +
Sbjct 508 PQYYIPKDPYPVPHYYPQQPLPLFDSSEMTELVDPDTLFYMFYYRPGTYQQYIAGQELKK 567
Query 78 RQWTYHKEWRKWFRAAEAPGV 98
+ W +HK++ WF+ E P +
Sbjct 568 QSWRFHKKYTTWFQRHEEPKM 588
> 7298223
Length=438
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/86 (33%), Positives = 41/86 (47%), Gaps = 2/86 (2%)
Query 13 FTSPISETPVT-KTVEELLPQCYVAAAPTPFRTSHLEKLETRTLFYIFYTSPRLLLQGYA 71
FT PI E +T + + LP Y+ + +E++ LF+ FYT ++Q A
Sbjct 296 FTGPIQEARLTPQEMSYKLPLNYLFNVNMSLQEPKIEEMHDELLFFFFYTYAGDMMQMLA 355
Query 72 AVELTRRQWTYHKEWRKW-FRAAEAP 96
A EL R W YHK W R A+ P
Sbjct 356 AAELAERGWRYHKYEHFWVIRQADNP 381
> ECU06g1510
Length=227
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 31/52 (59%), Gaps = 0/52 (0%)
Query 47 LEKLETRTLFYIFYTSPRLLLQGYAAVELTRRQWTYHKEWRKWFRAAEAPGV 98
+KL+ TLF+IFY+ + Q YAAV+L W +H ++ WF+ + P +
Sbjct 137 FKKLDIDTLFFIFYSQLGTIQQYYAAVQLKTYSWRFHTKYFTWFQRLDEPKL 188
> YDL165w
Length=191
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 54 TLFYIFYTSPRLLLQGYAAVELTRRQWTYHKEWRKW 89
TLF++FY P ++Q +EL +R W YHK + W
Sbjct 111 TLFFLFYKHPGTVIQELTYLELRKRNWRYHKTLKAW 146
> 7302229
Length=948
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 48 EKLETRTLFYIFYTSPRLLLQGYAAVELTRRQWTYHKEWRKWFRAAEAPGV 98
++L T TLF++FY Q AA L ++ W +H ++ WF+ E P +
Sbjct 859 QRLSTETLFFVFYYMEGSKAQYLAAKALKKQSWRFHTKYMMWFQRHEEPKI 909
> Hs7657387
Length=753
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 48 EKLETRTLFYIFYTSPRLLLQGYAAVELTRRQWTYHKEWRKWFRAAEAP 96
++L T TLF+IFY Q AA L ++ W +H ++ WF+ E P
Sbjct 664 QRLSTETLFFIFYYLEGTKAQYLAAKALKKQSWRFHTKYMMWFQRHEEP 712
> At5g18230
Length=889
Score = 37.4 bits (85), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 52 TRTLFYIFYTSPRLLLQGYAAVELTRRQWTYHKEWRKWFRAAEAPGVSS 100
T TLF+ FY Q AA EL ++ W YH+++ WF+ + P +++
Sbjct 795 TDTLFFAFYYQQNSYQQYLAAKELKKQSWRYHRKFNTWFQRHKEPKIAT 843
Lambda K H
0.315 0.128 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1888713112
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40