bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2022_orf2
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
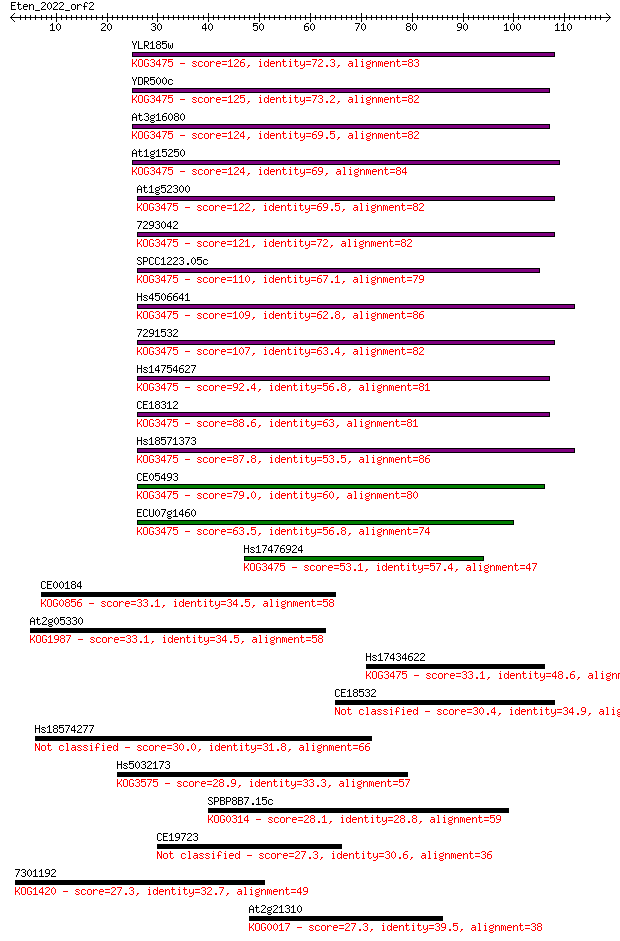
YLR185w 126 1e-29
YDR500c 125 1e-29
At3g16080 124 4e-29
At1g15250 124 4e-29
At1g52300 122 1e-28
7293042 121 2e-28
SPCC1223.05c 110 6e-25
Hs4506641 109 1e-24
7291532 107 4e-24
Hs14754627 92.4 2e-19
CE18312 88.6 3e-18
Hs18571373 87.8 5e-18
CE05493 79.0 2e-15
ECU07g1460 63.5 9e-11
Hs17476924 53.1 1e-07
CE00184 33.1 0.12
At2g05330 33.1 0.12
Hs17434622 33.1 0.13
CE18532 30.4 0.79
Hs18574277 30.0 1.1
Hs5032173 28.9 2.7
SPBP8B7.15c 28.1 4.0
CE19723 27.3 6.6
7301192 27.3 6.7
At2g21310 27.3 7.1
> YLR185w
Length=88
Score = 126 bits (316), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 60/83 (72%), Positives = 70/83 (84%), Gaps = 0/83 (0%)
Query 25 GKGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGT 84
GKGT SFG+RH K+H LC RCGRRS+HVQKKTC+SCGYP+AK R+YNW KAKRRHTTGT
Sbjct 2 GKGTPSFGKRHNKSHTLCNRCGRRSFHVQKKTCSSCGYPAAKTRSYNWGAKAKRRHTTGT 61
Query 85 GRMRYLKTLPRRAKNQFREGTVA 107
GRMRYLK + RR KN F+ G+ +
Sbjct 62 GRMRYLKHVSRRFKNGFQTGSAS 84
> YDR500c
Length=88
Score = 125 bits (315), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 60/82 (73%), Positives = 70/82 (85%), Gaps = 0/82 (0%)
Query 25 GKGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGT 84
GKGT SFG+RH K+H LC RCGRRS+HVQKKTC+SCGYPSAK R++NW+ KAKRRHTTGT
Sbjct 2 GKGTPSFGKRHNKSHTLCNRCGRRSFHVQKKTCSSCGYPSAKTRSHNWAAKAKRRHTTGT 61
Query 85 GRMRYLKTLPRRAKNQFREGTV 106
GRMRYLK + RR KN F+ G+
Sbjct 62 GRMRYLKHVSRRFKNGFQTGSA 83
> At3g16080
Length=142
Score = 124 bits (311), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 57/82 (69%), Positives = 68/82 (82%), Gaps = 0/82 (0%)
Query 25 GKGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGT 84
GKGTGSFG+R K+H LC+RCGRRS+H+QK C++C YP+A+KRTYNWS KA RR TTGT
Sbjct 2 GKGTGSFGKRRNKSHTLCVRCGRRSFHIQKSRCSACAYPAARKRTYNWSVKAIRRKTTGT 61
Query 85 GRMRYLKTLPRRAKNQFREGTV 106
GRMRYL+ +PRR K FREG V
Sbjct 62 GRMRYLRNVPRRFKTGFREGNV 83
> At1g15250
Length=95
Score = 124 bits (311), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 58/84 (69%), Positives = 69/84 (82%), Gaps = 0/84 (0%)
Query 25 GKGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGT 84
GKGTGSFG+R K+H LC+RCGRRS+H+QK C++C YP+A+KRTYNWS KA RR TTGT
Sbjct 2 GKGTGSFGKRRNKSHTLCVRCGRRSFHIQKSRCSACAYPAARKRTYNWSVKAIRRKTTGT 61
Query 85 GRMRYLKTLPRRAKNQFREGTVAV 108
GRMRYL+ +PRR K FREGT A
Sbjct 62 GRMRYLRNVPRRFKTCFREGTQAT 85
> At1g52300
Length=95
Score = 122 bits (307), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 57/82 (69%), Positives = 68/82 (82%), Gaps = 0/82 (0%)
Query 26 KGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTG 85
KGTGSFG+R K+H LC+RCGRRS+H+QK C++C YP+A+KRTYNWS KA RR TTGTG
Sbjct 3 KGTGSFGKRRNKSHTLCVRCGRRSFHIQKSRCSACAYPAARKRTYNWSVKAIRRKTTGTG 62
Query 86 RMRYLKTLPRRAKNQFREGTVA 107
RMRYL+ +PRR K FREGT A
Sbjct 63 RMRYLRNVPRRFKTGFREGTEA 84
> 7293042
Length=93
Score = 121 bits (304), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 59/82 (71%), Positives = 67/82 (81%), Gaps = 0/82 (0%)
Query 26 KGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTG 85
KGT SFG+RH KTH LC RCGR SYH+QK TCA CGYP+AK R+YNWS KAKRR TTGTG
Sbjct 3 KGTSSFGKRHNKTHTLCRRCGRSSYHIQKSTCAQCGYPAAKLRSYNWSVKAKRRKTTGTG 62
Query 86 RMRYLKTLPRRAKNQFREGTVA 107
RM++LK + RR +N FREGT A
Sbjct 63 RMQHLKVVRRRFRNGFREGTQA 84
> SPCC1223.05c
Length=91
Score = 110 bits (275), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 53/79 (67%), Positives = 61/79 (77%), Gaps = 0/79 (0%)
Query 26 KGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTG 85
KGT SFG RH K+H +C RCG+RS+H+QK TCA CGYP+AK R+YNW KAKRR TTGTG
Sbjct 3 KGTQSFGMRHNKSHTICRRCGKRSFHIQKSTCACCGYPAAKTRSYNWGAKAKRRRTTGTG 62
Query 86 RMRYLKTLPRRAKNQFREG 104
RM YLK + R KN FR G
Sbjct 63 RMSYLKKVHRSFKNGFRSG 81
> Hs4506641
Length=97
Score = 109 bits (273), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 54/86 (62%), Positives = 66/86 (76%), Gaps = 0/86 (0%)
Query 26 KGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTG 85
KGT SFG+R KTH LC RCG ++YH+QK TC CGYP+ +KR YNWS KAKRR+TTGTG
Sbjct 3 KGTSSFGKRRNKTHTLCRRCGSKAYHLQKSTCGKCGYPAKRKRKYNWSAKAKRRNTTGTG 62
Query 86 RMRYLKTLPRRAKNQFREGTVAVCQR 111
RMR+LK + RR ++ FREGT +R
Sbjct 63 RMRHLKIVYRRFRHGFREGTTPKPKR 88
> 7291532
Length=89
Score = 107 bits (268), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 52/82 (63%), Positives = 60/82 (73%), Gaps = 0/82 (0%)
Query 26 KGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTG 85
KGT SFG+RH KTH +C RCG SYH+QK C+ CGYP+AK R++NWS KAK R GTG
Sbjct 3 KGTTSFGKRHNKTHTICRRCGNSSYHLQKSKCSQCGYPAAKTRSFNWSRKAKGRKAQGTG 62
Query 86 RMRYLKTLPRRAKNQFREGTVA 107
RMRYLK L RR +N REG A
Sbjct 63 RMRYLKNLRRRFRNGLREGGAA 84
> Hs14754627
Length=97
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 46/81 (56%), Positives = 59/81 (72%), Gaps = 0/81 (0%)
Query 26 KGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTG 85
KGT FG+R KTH LC R G ++YH+QK TC CGY + +KR YNWS KAKR++TTGTG
Sbjct 3 KGTSLFGKRRNKTHTLCCRRGSKAYHLQKSTCGKCGYAAKRKRKYNWSAKAKRQNTTGTG 62
Query 86 RMRYLKTLPRRAKNQFREGTV 106
MR+LK + R ++ F EGT+
Sbjct 63 GMRHLKIVYHRFRHGFYEGTI 83
> CE18312
Length=92
Score = 88.6 bits (218), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 51/81 (62%), Positives = 63/81 (77%), Gaps = 0/81 (0%)
Query 26 KGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTG 85
KGT +FG++H K+H LC RCG+ S+H+QKK CASCGYP AKKRTYNW K+ RR TTGTG
Sbjct 3 KGTQAFGKKHVKSHTLCKRCGKSSFHIQKKRCASCGYPDAKKRTYNWGAKSIRRRTTGTG 62
Query 86 RMRYLKTLPRRAKNQFREGTV 106
R R+L+ + R +N FREGT
Sbjct 63 RTRHLRDVNARFRNGFREGTT 83
> Hs18571373
Length=97
Score = 87.8 bits (216), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 46/86 (53%), Positives = 60/86 (69%), Gaps = 0/86 (0%)
Query 26 KGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTG 85
KGT SFG+ KT+ LC CG ++YH+QK T CGY + +KR YNWS KAKRR+TT TG
Sbjct 3 KGTSSFGKHCNKTYTLCHHCGSKAYHLQKSTYGKCGYTAKRKRKYNWSTKAKRRNTTRTG 62
Query 86 RMRYLKTLPRRAKNQFREGTVAVCQR 111
+MR+LK + R ++ FREGT +R
Sbjct 63 QMRHLKIVYHRFRHGFREGTTPKPKR 88
> CE05493
Length=91
Score = 79.0 bits (193), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 48/80 (60%), Positives = 60/80 (75%), Gaps = 0/80 (0%)
Query 26 KGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTG 85
KGT +FG++H K+H LC RCG+ S+H+QKK CASCGY AKKRTYNW K+ RR TTGTG
Sbjct 3 KGTQAFGKKHVKSHTLCKRCGKSSFHIQKKRCASCGYQDAKKRTYNWGAKSIRRRTTGTG 62
Query 86 RMRYLKTLPRRAKNQFREGT 105
R R+L+ + R +N FR T
Sbjct 63 RTRHLRDVNARFRNGFRGTT 82
> ECU07g1460
Length=90
Score = 63.5 bits (153), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 42/74 (56%), Positives = 53/74 (71%), Gaps = 1/74 (1%)
Query 26 KGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTG 85
KGT SFG+++ + +C RCGR+SYH QK +C+SCGYP+ K R S KA+RR T GTG
Sbjct 3 KGTASFGKKNKRNTEMCRRCGRQSYHKQKNSCSSCGYPNPKMRNPG-SIKARRRRTIGTG 61
Query 86 RMRYLKTLPRRAKN 99
RMRY+K R AKN
Sbjct 62 RMRYMKRELRAAKN 75
> Hs17476924
Length=120
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/49 (55%), Positives = 35/49 (71%), Gaps = 2/49 (4%)
Query 47 RRSYHV--QKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTGRMRYLKTL 93
RRS+ + Q TC GYP+ +KR NW KAKRR++TGTGRMR+LK +
Sbjct 66 RRSFALREQMSTCGKRGYPAKRKRKCNWGAKAKRRNSTGTGRMRHLKIV 114
> CE00184
Length=152
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 28/59 (47%), Gaps = 5/59 (8%)
Query 7 LPNFVDFQVY-VHSKMGKAGKGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPS 64
LPN +VY V + G TG F K ++CL CG ++ K A CG+P+
Sbjct 34 LPN----EVYRVARESGTETPHTGGFNDHFEKGRYVCLCCGSELFNSDAKFWAGCGWPA 88
> At2g05330
Length=215
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Query 5 LGLPNFVDFQVYVHSKMGKAGKGTGSFGRRHGKTHFLCLRCGRRSYHVQKKT-CASCGY 62
L P+F +F+++V + A + + R T C RCG +YH Q T C SCG+
Sbjct 153 LMFPSFDEFKLFVDNYPNLAVEVMMASLTRTPSTS--CSRCGLITYHNQTGTSCCSCGF 209
> Hs17434622
Length=170
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 71 NWSEKAKRRHTTGTGRMRYLKTLPRRAKNQFREGT 105
N S K KR++TTG +MRYLK + R +N+ EG
Sbjct 122 NCSAKVKRQNTTGICQMRYLKIVYWRFRNRVCEGI 156
> CE18532
Length=404
Score = 30.4 bits (67), Expect = 0.79, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 65 AKKRTYNWSEKAKRRHTTGTGRMRYLKTLPRRAKNQFREGTVA 107
+ K NW E + R+ T T M+ LKT P+ K+ TV+
Sbjct 68 SNKIDLNWGEGGESRYVTITRLMKILKTSPQSIKDLLPHNTVS 110
> Hs18574277
Length=740
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 30/69 (43%), Gaps = 4/69 (5%)
Query 6 GLP---NFVDFQVYVHSKMGKAGKGTGSFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGY 62
GLP N V F V+V K TG GR H ++ + C R+ H+ T C
Sbjct 190 GLPIIINAVVFAVFVQGKTTLWKNITGKGGRDHKRSQDVLPDCVLRNAHIPGSTLLICS- 248
Query 63 PSAKKRTYN 71
P+ + Y+
Sbjct 249 PAQEALAYS 257
> Hs5032173
Length=423
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query 22 GKAG-KGTGSFGRRHGKTHFLCLRCGRRSY--HVQKKTCASCG--YPSAKKRTYNWSEKA 76
G AG KG G G + H C+ CG +S H + TC C + + +R ++ +A
Sbjct 65 GTAGDKGQGPPGSGQSQQHIECVVCGDKSSGKHYGQFTCEGCKSFFKRSVRRNLTYTCRA 124
Query 77 KR 78
R
Sbjct 125 NR 126
> SPBP8B7.15c
Length=482
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 30/59 (50%), Gaps = 10/59 (16%)
Query 40 FLCLRCGRRSYHVQKKTCASCGYPSAKKRTYNWSEKAKRRHTTGTGRMRYLKTLPRRAK 98
++C RCG++ + +Q C + P N+ K + + TTG R +LK + R A+
Sbjct 183 YICYRCGQKGHWIQ--ACPTNADP-------NYDGKPRVKRTTGIPR-SFLKNVERPAE 231
> CE19723
Length=671
Score = 27.3 bits (59), Expect = 6.6, Method: Composition-based stats.
Identities = 11/36 (30%), Positives = 20/36 (55%), Gaps = 2/36 (5%)
Query 30 SFGRRHGKTHFLCLRCGRRSYHVQKKTCASCGYPSA 65
++ + + TH C +CG+ + + +K C CG P A
Sbjct 93 TYNKNNFSTHHFCNKCGKVAQNSKK--CKHCGGPVA 126
> 7301192
Length=1175
Score = 27.3 bits (59), Expect = 6.7, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 21/49 (42%), Gaps = 2/49 (4%)
Query 2 LLLLGLPNFVDFQVYVHSKMGKAGKGTGSFGRRHGKTHFLCLRCGRRSY 50
LL+GL F + +G K G R HGK H + CG +Y
Sbjct 321 FLLVGLAMFASSIPEIIELVGSGNKYGGELKREHGKRHIVV--CGHITY 367
> At2g21310
Length=838
Score = 27.3 bits (59), Expect = 7.1, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 20/39 (51%), Gaps = 1/39 (2%)
Query 48 RSYHVQKKTCASCGYPSA-KKRTYNWSEKAKRRHTTGTG 85
RS +KK C CG KK+ Y W EK K+ + + G
Sbjct 235 RSKSREKKVCWVCGKEGHFKKQCYVWKEKNKKGNNSEKG 273
Lambda K H
0.323 0.135 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40