bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2008_orf1
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
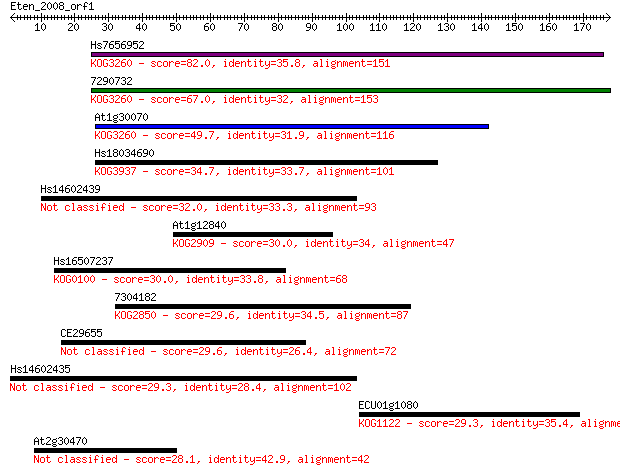
Hs7656952 82.0 5e-16
7290732 67.0 2e-11
At1g30070 49.7 3e-06
Hs18034690 34.7 0.11
Hs14602439 32.0 0.73
At1g12840 30.0 2.6
Hs16507237 30.0 2.8
7304182 29.6 3.3
CE29655 29.6 3.4
Hs14602435 29.3 4.2
ECU01g1080 29.3 4.7
At2g30470 28.1 8.9
> Hs7656952
Length=228
Score = 82.0 bits (201), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 54/163 (33%), Positives = 81/163 (49%), Gaps = 12/163 (7%)
Query 25 STEALRCDLEEFEAFLKLAKRPSVKSFLSAHCQRLRNQIE---------EAERLD--KCA 73
++E L+ DLEE + L+ A R V+ L+A ++ +I+ +AE LD K A
Sbjct 2 ASEELQKDLEEVKVLLEKATRKRVRDALTAEKSKIETEIKNKMQQKSQKKAELLDNEKPA 61
Query 74 SQVTPTVEVFNT-IDRFAWGQTRDLVKVYVQLDGLGSLPEEATSVHFEETSAQLVVQNLR 132
+ V P + I + W Q+ VK+Y+ L G+ +P E VHF E S L+V+NL
Sbjct 62 AVVAPITTGYTVKISNYGWDQSDKFVKIYITLTGVHQVPTENVQVHFTERSFDLLVKNLN 121
Query 133 HKNYILKLCPLYGPIDVGKSSFSRKRDTATLRLAKRRPGEHWD 175
K+Y + + L PI V SS K DT + K+ WD
Sbjct 122 GKSYSMIVNNLLKPISVEGSSKKVKTDTVLILCRKKVENTRWD 164
> 7290732
Length=230
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 49/158 (31%), Positives = 76/158 (48%), Gaps = 6/158 (3%)
Query 25 STEALRCDLEEFEAFLKLAKRPSVKSFLSAHCQRLRNQIEEAERLDKCAS--QVTPTVEV 82
S E L+ D+ E AFL+ AK VK L+ +I E K A+ Q T + E
Sbjct 2 SLEQLKSDVAELAAFLQQAKGARVKDVLTTAKAEAEREIVNLELKAKIAAERQATGSSEA 61
Query 83 ---FNTIDRFAWGQTRDLVKVYVQLDGLGSLPEEATSVHFEETSAQLVVQNLRHKNYILK 139
+ + + W Q+ VK+++ L+G+ EE +V + S QL V++L+ K++ L
Sbjct 62 KRYLHELTDYGWDQSAKFVKLFITLNGVQGCTEENVTVTYTPNSLQLHVRDLQGKDFGLT 121
Query 140 LCPLYGPIDVGKSSFSRKRDTATLRLAKRRPGEHWDTI 177
+ L IDV KS K D + L K +HWD +
Sbjct 122 VNNLLHSIDVEKSYRKIKTDMVAIYLQKVED-KHWDVL 158
> At1g30070
Length=222
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 37/138 (26%), Positives = 58/138 (42%), Gaps = 30/138 (21%)
Query 26 TEALRCDLEEFEAFLKLAKRPSVKSFLSAHCQRLRNQIEEAERLDKCASQVTPTVEV--- 82
E + DLEE +AKRP V + +++ L E R +S P V V
Sbjct 2 AEEVGLDLEELRQLQNIAKRPRVLNLINSEISNL-----EKLRDSAVSSNAKPKVPVTVP 56
Query 83 ----------------FNTIDRFAWGQTRDLVKVYVQLDGLGSLPEEATSVHFEETSAQL 126
+ T+ F+W Q D VK+Y+ L+G+ E+ F+ S +
Sbjct 57 APVSSSGKPVSSSALNYVTLGTFSWDQDNDKVKMYISLEGVD---EDKVQAEFKPMSLDI 113
Query 127 VVQNLRHKNY---ILKLC 141
+ +++ KNY I KLC
Sbjct 114 KIHDVQGKNYRCAIPKLC 131
> Hs18034690
Length=384
Score = 34.7 bits (78), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 34/113 (30%), Positives = 51/113 (45%), Gaps = 17/113 (15%)
Query 26 TEALRCDLEEFEAFLKLAKRPSVKSFLSAHCQRLRNQIEEA--ERLDK-----CA-SQVT 77
EA+R +L+E + FL ++K ++S L N I EA E+L CA S V
Sbjct 108 VEAMRANLQELDQFLGPYPYATLKKWIS-----LTNFISEATVEKLQPENRQICAFSDVL 162
Query 78 PTVEVFNTIDRFAWGQTRDLVKVYVQLDGLGSLPE----EATSVHFEETSAQL 126
P + + +T DR R ++ +GL LPE T + F E Q+
Sbjct 163 PVLSMKHTKDRVGQNLPRCGIECKSYQEGLARLPEMKPRAGTEIRFSELPTQM 215
> Hs14602439
Length=831
Score = 32.0 bits (71), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 45/101 (44%), Gaps = 10/101 (9%)
Query 10 RTRRISFKDRRLTMDSTEA-----LRCDLEEFEAFLK-LAKRPSVK--SFLSAHCQRLRN 61
R R S K+RR+T S E ++ + + L+ +PS+K + L Q+LR+
Sbjct 639 RGRPDSNKNRRITHISAEQKRRFNIKLGFDTLHGLVSTLSAQPSLKERAGLQEEAQQLRD 698
Query 62 QIEEAERLDKCASQVTPTVEVFNTIDRFAWGQTRDLVKVYV 102
+IEE Q P V T RF Q RD+ YV
Sbjct 699 EIEELNAAINLCQQQLPATGVPITHQRF--DQMRDMFDDYV 737
> At1g12840
Length=370
Score = 30.0 bits (66), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 24/49 (48%), Gaps = 2/49 (4%)
Query 49 KSFLSAHCQRLRNQIEEAERLDKCASQVTPT--VEVFNTIDRFAWGQTR 95
SF+ Q++R QIEE ER+ S V V + + RF W + +
Sbjct 63 NSFVEGVSQKIRRQIEELERISGVESNALTVDGVPVDSYLTRFVWDEAK 111
> Hs16507237
Length=654
Score = 30.0 bits (66), Expect = 2.8, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 38/70 (54%), Gaps = 2/70 (2%)
Query 14 ISFKDRRLTMDSTEALRCDLEEF-EAFLKLAKRPSVKSFLSAHCQRLRNQIEEAERL-DK 71
I+ RLT + E + D E+F E KL +R ++ L ++ L+NQI + E+L K
Sbjct 526 ITNDQNRLTPEEIERMVNDAEKFAEEDKKLKERIDTRNELESYAYSLKNQIGDKEKLGGK 585
Query 72 CASQVTPTVE 81
+S+ T+E
Sbjct 586 LSSEDKETME 595
> 7304182
Length=271
Score = 29.6 bits (65), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 44/94 (46%), Gaps = 8/94 (8%)
Query 32 DLEEFEAFLKLAKRPSVKSFLSAHCQRLRNQIEEAERLDKCASQVTPTV---EVFNTIDR 88
D +EFE L LA+RP+ + L + L +++E + L A + +V + N IDR
Sbjct 30 DDDEFEDLLPLARRPNGHARLGRYENTLEVKVQEGDTLQALALRFHSSVADIKRLNKIDR 89
Query 89 ----FAWGQTRDLVKVYVQLDGLGSLPEEATSVH 118
A R V V+ L G G L E +VH
Sbjct 90 ENEIHAHRVIRIPVTVHNVLLGNGGL-EALPAVH 122
> CE29655
Length=7659
Score = 29.6 bits (65), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 35/73 (47%), Gaps = 3/73 (4%)
Query 16 FKDRRLTMDSTEALRCDLEEF-EAFLKLAKRPSVKSFLSAHCQRLRNQIEEAERLDKCAS 74
K++ +D L+ D ++ EA L L F+ H + + Q E+ +DK +
Sbjct 7358 LKEQSYLLDRIRDLKIDFDDLGEALLPLTVAEDELRFMHVHVESIERQYEDT--MDKLNA 7415
Query 75 QVTPTVEVFNTID 87
++T VE+ T+D
Sbjct 7416 EITAEVELLRTLD 7428
> Hs14602435
Length=833
Score = 29.3 bits (64), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 29/104 (27%), Positives = 45/104 (43%), Gaps = 16/104 (15%)
Query 1 KKVQYIHEKRTRRISFKDRRLTMDSTEALRCDLEEFEAFLKLAKRPSVK--SFLSAHCQR 58
+++ +I ++ RR + K L D+ L L+ +PS+K + L Q+
Sbjct 650 RRITHISAEQKRRFNIK---LGFDTLHGLVS---------TLSAQPSLKERAGLQEEAQQ 697
Query 59 LRNQIEEAERLDKCASQVTPTVEVFNTIDRFAWGQTRDLVKVYV 102
LR++IEE Q P V T RF Q RD+ YV
Sbjct 698 LRDEIEELNAAINLCQQQLPATGVPITHQRF--DQMRDMFDDYV 739
> ECU01g1080
Length=370
Score = 29.3 bits (64), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 33/72 (45%), Gaps = 9/72 (12%)
Query 104 LDGLGSLPEEATSVHFEETSAQLVVQNLRHKNYILKLCP----LYGPIDVGKS---SFSR 156
L G G L + + T + LV +N NY+L CP ID+GK SF
Sbjct 281 LHGFGML--KPGGILMYSTCSVLVKENEEVVNYLLSKCPSAKMAECEIDIGKDGFMSFKG 338
Query 157 KRDTATLRLAKR 168
K +L++A+R
Sbjct 339 KNYNGSLKMARR 350
> At2g30470
Length=780
Score = 28.1 bits (61), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 8 EKRTRRISFKDRRLTMDSTEALRCDLEEFEAFLKLAKRPSVK 49
+KRTR I K++RL + S E++ L EA L PSVK
Sbjct 456 KKRTRTIGAKNKRLLLHSEESMELRLTWEEAQDLLRPSPSVK 497
Lambda K H
0.321 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2743263016
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40