bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2005_orf1
Length=187
Score E
Sequences producing significant alignments: (Bits) Value
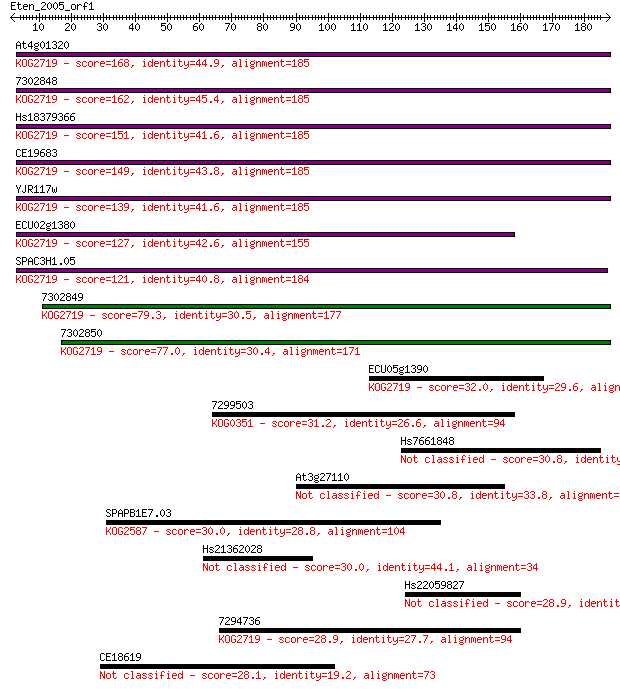
At4g01320 168 7e-42
7302848 162 3e-40
Hs18379366 151 7e-37
CE19683 149 2e-36
YJR117w 139 4e-33
ECU02g1380 127 1e-29
SPAC3H1.05 121 8e-28
7302849 79.3 4e-15
7302850 77.0 2e-14
ECU05g1390 32.0 0.72
7299503 31.2 1.1
Hs7661848 30.8 1.4
At3g27110 30.8 1.7
SPAPB1E7.03 30.0 2.7
Hs21362028 30.0 2.7
Hs22059827 28.9 6.4
7294736 28.9 6.4
CE18619 28.1 10.0
> At4g01320
Length=459
Score = 168 bits (425), Expect = 7e-42, Method: Compositional matrix adjust.
Identities = 83/187 (44%), Positives = 122/187 (65%), Gaps = 2/187 (1%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FNK+T +FI+D + G L+ +G PI+ A+ ++++ GG +LW F + + M+ I
Sbjct 147 FNKQTIWMFIRDMIKGTFLSVILGPPIVAAIIFIVQKGGPYLAIYLWAFMFILSLVMMTI 206
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
YP IAPLFNKF L D +LR KI+ LA L FPL +++ +DGS RSSHSNAY YGF++
Sbjct 207 YPVLIAPLFNKFTPLPDGDLREKIEKLASSLKFPLKKLFVVDGSTRSSHSNAYMYGFFKN 266
Query 123 KRIVIFDTLLHL--PHDQILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNND 180
KRIV++DTL+ D+I+A++ HELGHW++NH + V + F+ F Y V N+
Sbjct 267 KRIVLYDTLIQQCKNEDEIVAVIAHELGHWKLNHTTYSFIAVQILAFLQFGGYTLVRNST 326
Query 181 HLYKCFG 187
L++ FG
Sbjct 327 DLFRSFG 333
> 7302848
Length=451
Score = 162 bits (410), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 84/203 (41%), Positives = 124/203 (61%), Gaps = 18/203 (8%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FNK+TA F D+L G +T + PI A+ ++++ GG+NF WLW F+ + + ++ +
Sbjct 146 FNKQTARFFAWDQLKGFLVTQVLMIPITAAIIFIVQRGGDNFFIWLWIFTGVISLVLLTL 205
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
YP FIAPLF+K+ L LR I+ LA L FPL +++ ++GSKRSSHSNAYFYG W
Sbjct 206 YPIFIAPLFDKYTPLEKGALRQSIEDLAASLKFPLTKLFVVEGSKRSSHSNAYFYGLWNS 265
Query 123 KRIVIFDTLL---HLPHD---------------QILAILGHELGHWQMNHFMQRLLLVFM 164
KRIV+FDTLL P D ++LA+LGHELGHW++ H + ++++ +
Sbjct 266 KRIVLFDTLLLNKGKPDDSELSEEEKGKGCTDEEVLAVLGHELGHWKLGHVTKNIIIMQV 325
Query 165 NLFVTFYLYGKVMNNDHLYKCFG 187
+LF+ F ++G V Y G
Sbjct 326 HLFLMFLVFGNVFKYPPFYVAMG 348
> Hs18379366
Length=475
Score = 151 bits (382), Expect = 7e-37, Method: Compositional matrix adjust.
Identities = 77/223 (34%), Positives = 130/223 (58%), Gaps = 38/223 (17%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FN++T F+KD + +T I P+ + ++IK GG+ F + W F+++ + ++ I
Sbjct 155 FNQQTLGFFMKDAIKKFVVTQCILLPVSSLLLYIIKIGGDYFFIYAWLFTLVVSLVLVTI 214
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
Y ++IAPLF+KF L + +L+ +I+ +A+ ++FPL +VY ++GSKRSSHSNAYFYGF++
Sbjct 215 YADYIAPLFDKFTPLPEGKLKEEIEVMAKSIDFPLTKVYVVEGSKRSSHSNAYFYGFFKN 274
Query 123 KRIVIFDTLLH--------------------------------------LPHDQILAILG 144
KRIV+FDTLL ++++LA+LG
Sbjct 275 KRIVLFDTLLEEYSVLNKDIQEDSGMEPRNEEEGNSEEIKAKVKNKKQGCKNEEVLAVLG 334
Query 145 HELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNNDHLYKCFG 187
HELGHW++ H ++ +++ MN F+ F+L+ ++ L+ FG
Sbjct 335 HELGHWKLGHTVKNIIISQMNSFLCFFLFAVLIGRKELFAAFG 377
> CE19683
Length=442
Score = 149 bits (377), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 81/213 (38%), Positives = 126/213 (59%), Gaps = 28/213 (13%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FNK+T + DK+ + + A+ PI+ + W+I GG F ++W F + ++ I
Sbjct 131 FNKQTIGFYFVDKIKKMLVGFALTMPIVYGIEWIIVNGGPYFFVYIWLFVSVVVLLLMTI 190
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
YP FIAPLF+K+ L D +L++KI+ LA L++PL E+Y ++GSKRS+HSNAY YGFW+
Sbjct 191 YPTFIAPLFDKYFPLPDGDLKTKIEQLAASLSYPLTELYVVNGSKRSAHSNAYMYGFWKN 250
Query 123 KRIVIFDTLLH----------------------------LPHDQILAILGHELGHWQMNH 154
KRIV++DTLL + +D+++A+LGHELGHW + H
Sbjct 251 KRIVLYDTLLSGAEKEKVHELYVAAGEKIEETENDKKRGMNNDEVVAVLGHELGHWALWH 310
Query 155 FMQRLLLVFMNLFVTFYLYGKVMNNDHLYKCFG 187
+ L++ +NLF +F ++G + LY+ FG
Sbjct 311 TLINLVITEVNLFFSFAVFGYFYKWEALYQGFG 343
> YJR117w
Length=453
Score = 139 bits (350), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 77/187 (41%), Positives = 112/187 (59%), Gaps = 2/187 (1%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FNK T L+I D + L L AIG PIL + +F +++ F + + I
Sbjct 153 FNKLTVQLWITDMIKSLTLAYAIGGPILYLFLKIFDKFPTDFLWYIMVFLFVVQILAMTI 212
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGF-WR 121
P FI P+FNKF L D EL+ I++LA+++ FPL +++ +DGSKRSSHSNAYF G +
Sbjct 213 IPVFIMPMFNKFTPLEDGELKKSIESLADRVGFPLDKIFVIDGSKRSSHSNAYFTGLPFT 272
Query 122 WKRIVIFDTLLHL-PHDQILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNND 180
KRIV+FDTL++ D+I A+L HE+GHWQ NH + ++ ++ F+ F L+ + N
Sbjct 273 SKRIVLFDTLVNSNSTDEITAVLAHEIGHWQKNHIVNMVIFSQLHTFLIFSLFTSIYRNT 332
Query 181 HLYKCFG 187
Y FG
Sbjct 333 SFYNTFG 339
> ECU02g1380
Length=410
Score = 127 bits (318), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 66/157 (42%), Positives = 98/157 (62%), Gaps = 2/157 (1%)
Query 3 FNKKTAILFIKDKL-LGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIV 61
FNK T F+ D L + L +T G + N + K+ +F+ +LW F + G+++
Sbjct 126 FNKTTLSTFLMDFLKMSLIITVLFGPFSYVSTNIIKKYYKTSFYIYLWVFMAVFQIGLVI 185
Query 62 IYPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWR 121
+YP I PLFNKF+ + + L++KI+ LAE++ ++ MD SKRS HSNAYF G +
Sbjct 186 VYPIAIQPLFNKFEEMEESNLKTKIEKLAERVGICAKKILVMDASKRSGHSNAYFIGLTK 245
Query 122 WKRIVIFDTLL-HLPHDQILAILGHELGHWQMNHFMQ 157
KRIVI+DTLL + ++ LAIL HELGHW+ NH ++
Sbjct 246 EKRIVIYDTLLKQVDEEETLAILCHELGHWKHNHVVK 282
> SPAC3H1.05
Length=474
Score = 121 bits (304), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 75/188 (39%), Positives = 116/188 (61%), Gaps = 7/188 (3%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIV- 61
FNK T +F+ D L L L + + ++ ++ G+NF + WG ++ FG+I+
Sbjct 189 FNKSTLKIFVIDLLKELSLGGLLMSVVVGVFVKILTKFGDNFIMYAWGAYIV--FGLILQ 246
Query 62 -IYPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFW 120
I P+ I PLF KF L + LR++I+ LA +NFPL ++Y +D S+RS+HSNA+FYG
Sbjct 247 TIAPSLIMPLFYKFTPLENGSLRTQIEELAASINFPLKKLYVIDASRRSTHSNAFFYGLP 306
Query 121 RWKRIVIFDTLL-HLPHDQILAILGHELGHWQMNHFMQRLLLVF-MNLFVTFYLYGKVMN 178
K IV+FDTL+ + +++AILGHELGHW M+H + ++ + M+LF F L+ +
Sbjct 307 WNKGIVLFDTLVKNHTEPELIAILGHELGHWYMSHNLINTIIDYGMSLFHLF-LFAAFIR 365
Query 179 NDHLYKCF 186
N+ LY F
Sbjct 366 NNSLYTSF 373
> 7302849
Length=447
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 54/195 (27%), Positives = 100/195 (51%), Gaps = 22/195 (11%)
Query 11 FIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVIYPNFIAPL 70
F+ D ++G A I ++ A+ ++ G L+ S++ T ++++ P I P
Sbjct 159 FVVDLVVG----AMITTLVVVALVYMFIGLGPYAPLGLYLQSLILTMIVLLLIPFMIHPF 214
Query 71 FNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRWKRIVIFDT 130
+ L + LR++++ L Q+ FP+ +V + ++ SNA+FYG KRIVIFDT
Sbjct 215 VGQSVPLENSNLRTQLEYLTRQVGFPMSQVRIIRVHDPNTGSNAFFYGCCCLKRIVIFDT 274
Query 131 LL---------HLPHD---------QILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYL 172
LL HL + Q++A++ HELGHW+ HF + ++ ++L +T L
Sbjct 275 LLLNRGKSDLSHLTAEELGRGLADPQVVAVVAHELGHWRNGHFYKAIMAFQVHLILTILL 334
Query 173 YGKVMNNDHLYKCFG 187
+ + ++ +Y+ G
Sbjct 335 FAFLFSHGPIYQAVG 349
> 7302850
Length=426
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 52/189 (27%), Positives = 91/189 (48%), Gaps = 19/189 (10%)
Query 17 LGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVIYPNFIAPLFNKFKV 76
+ + L+ + P+ A+ + +K+ G F W W F T ++ P P + V
Sbjct 133 MSILLSQLVLFPLAAAIVFSVKFIGYYFFLWFWLFWATFTLLLVFFLPYCCIPCIGRQVV 192
Query 77 LGD-RELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRWKRIVIFDTLL--- 132
L + L ++ + + + FP+ V+ + ++ +SNAYFYG KRIVIFDTLL
Sbjct 193 LPEGTALYMEVKRVCDVVGFPMKRVFIIK-TRTMQYSNAYFYGSCCLKRIVIFDTLLLNK 251
Query 133 --------------HLPHDQILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMN 178
L + Q+ ++ HELGHW+ HF + +++ ++ F+T L+G +
Sbjct 252 GKEPNEIHPYEVGRGLTNIQVAGVVCHELGHWKHGHFYKATIIMKIHFFITMGLFGLFFH 311
Query 179 NDHLYKCFG 187
+ LY G
Sbjct 312 SPQLYMAVG 320
> ECU05g1390
Length=404
Score = 32.0 bits (71), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 113 NAYFYGFWRWKRIVIFDTLLHLPHDQILAILGHELGHWQMNHFMQRLLLVFMNL 166
NA G +RI I+ + + ++Q+ +IL HE+GH M+++ + F+ L
Sbjct 239 NAALIGIGSAERIEIYGKVEKIDNEQLESILIHEVGHSCHRSLMKKISVFFIIL 292
> 7299503
Length=1487
Score = 31.2 bits (69), Expect = 1.1, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 46/94 (48%), Gaps = 7/94 (7%)
Query 64 PNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRWK 123
P+ APL ++ V D L + LA+ NF +D +K+++ + GF+R
Sbjct 327 PSPAAPLKPRYSVAFDNSLADYLKDLAQNDNF------SIDPNKQNTETLKSTLGFFRNT 380
Query 124 RIVIFDTLLHLPHDQILAILGHELGHWQMNHFMQ 157
+ + + L DQI A+ +E+ +Q N F++
Sbjct 381 YVELMEKYCSLI-DQIPAMHFNEIAGFQPNTFLK 413
> Hs7661848
Length=338
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 38/65 (58%), Gaps = 6/65 (9%)
Query 123 KRIVIFDTLLHLPHDQILAILGHE-LGHWQMNHFMQRL--LLVFMNLFVTFYLYGKVMNN 179
K IVI D ++ L ILG LG WQ+N F+ R+ +L ++N++V+ +G +++
Sbjct 61 KNIVIADFVMSLTFP--FKILGDSGLGPWQLNVFVCRVSAVLFYVNMYVSIVFFG-LISF 117
Query 180 DHLYK 184
D YK
Sbjct 118 DRYYK 122
> At3g27110
Length=344
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 34/67 (50%), Gaps = 6/67 (8%)
Query 90 AEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRWKR-IVIFDTLLHL-PHDQILAILGHEL 147
AE LN ++Y ++S NAY K IV+ +L+ L ++ A+L HEL
Sbjct 138 AEILNIEAPDLY----VRQSPVPNAYTLAISGKKPFIVVHTSLIELLTSAELQAVLAHEL 193
Query 148 GHWQMNH 154
GH + +H
Sbjct 194 GHLKCDH 200
> SPAPB1E7.03
Length=591
Score = 30.0 bits (66), Expect = 2.7, Method: Composition-based stats.
Identities = 30/128 (23%), Positives = 49/128 (38%), Gaps = 27/128 (21%)
Query 31 CAVNWLIKWGGENFHFWLWGFSVLTTFGMIVIYPNFIAPLFNK----------------- 73
C++ ++ K G W F+ LT + Y NFI F +
Sbjct 396 CSLKFISKIGNRGMGEWAVNFTHLTDMLRAIEYENFIEQKFGERAIRLLRIIKDKGKIEE 455
Query 74 -----FKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRWKRI--V 126
+L R+LR+ + A+AE E+ E+ S + S +F F R R +
Sbjct 456 KQLANIALLRQRDLRTVLQAMAE---IGALELQEVPRSSDRAPSKTFFLWFHRPDRAYSL 512
Query 127 IFDTLLHL 134
+ D L H+
Sbjct 513 LLDELYHV 520
> Hs21362028
Length=760
Score = 30.0 bits (66), Expect = 2.7, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 61 VIYPNFIAPLFNKFKVLGDRELRSKIDALAEQLN 94
VI NF+ P +NK +E R I+ALA++LN
Sbjct 187 VIECNFLKPAYNKQDFEYTKEYRLTINALAQKLN 220
> Hs22059827
Length=156
Score = 28.9 bits (63), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 11/36 (30%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 124 RIVIFDTLLHLPHDQILAILGHELGHWQMNHFMQRL 159
++ +F +LH PH ++L++ G L H ++H + L
Sbjct 51 KVELFKAVLHDPHLKLLSLYGTSLSHTDVSHLCETL 86
> 7294736
Length=504
Score = 28.9 bits (63), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 26/111 (23%), Positives = 43/111 (38%), Gaps = 19/111 (17%)
Query 66 FIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRWKRI 125
F P K + + ++ + + A+ + NFP VY M + A+ G R+
Sbjct 264 FGVPCLGKSRKMNSTDMDNSLKAVLDDFNFP-GRVY-MVHTFHVGRPTAWVMGCCCCLRL 321
Query 126 VIFDTL-----------------LHLPHDQILAILGHELGHWQMNHFMQRL 159
I D L L +Q+ A + H+L HWQ+ H + L
Sbjct 322 DIHDNLKWNRGFSSDDFDWGQMGAGLNDEQLAAFVAHQLAHWQLWHVAKGL 372
> CE18619
Length=338
Score = 28.1 bits (61), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 14/78 (17%), Positives = 34/78 (43%), Gaps = 5/78 (6%)
Query 29 ILCAVNWLIKWGG-----ENFHFWLWGFSVLTTFGMIVIYPNFIAPLFNKFKVLGDRELR 83
++C +W KW FH++++ F F ++ + + +F F L
Sbjct 125 VICTFSWKPKWESFKSMFMPFHYFIYTFMFSIIFFLVPDQKSALQQVFQTFPCLPREIYE 184
Query 84 SKIDALAEQLNFPLCEVY 101
+ + +A+ + FP+ ++
Sbjct 185 APVYVIADDVTFPVIVIF 202
Lambda K H
0.331 0.145 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3058524910
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40