bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1971_orf2
Length=272
Score E
Sequences producing significant alignments: (Bits) Value
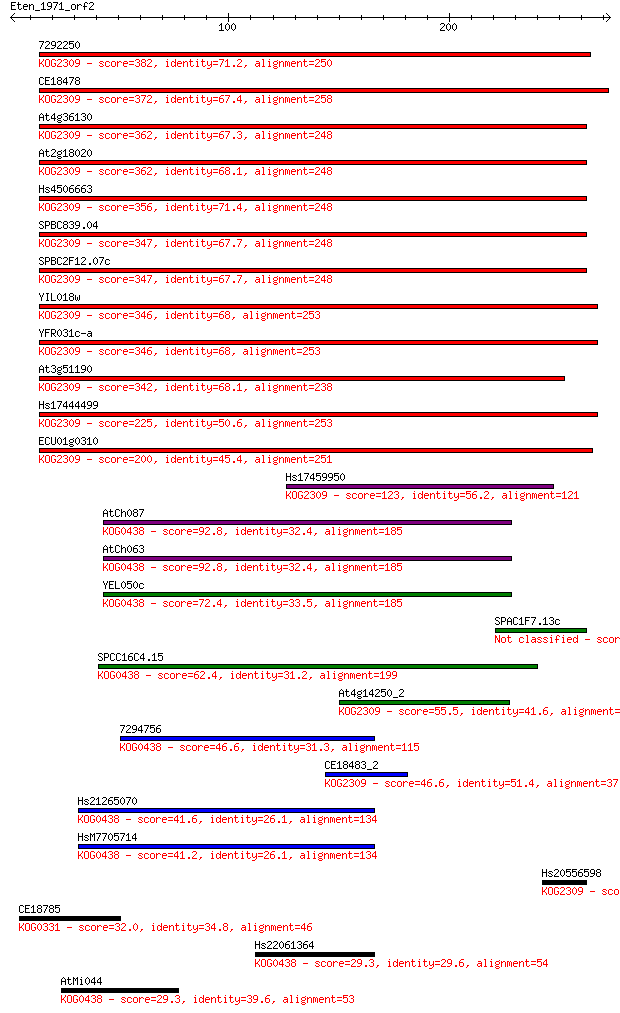
7292250 382 5e-106
CE18478 372 5e-103
At4g36130 362 5e-100
At2g18020 362 6e-100
Hs4506663 356 3e-98
SPBC839.04 347 1e-95
SPBC2F12.07c 347 1e-95
YIL018w 346 3e-95
YFR031c-a 346 3e-95
At3g51190 342 7e-94
Hs17444499 225 9e-59
ECU01g0310 200 3e-51
Hs17459950 123 5e-28
AtCh087 92.8 6e-19
AtCh063 92.8 6e-19
YEL050c 72.4 8e-13
SPAC1F7.13c 72.0 1e-12
SPCC16C4.15 62.4 1e-09
At4g14250_2 55.5 1e-07
7294756 46.6 6e-05
CE18483_2 46.6 6e-05
Hs21265070 41.6 0.002
HsM7705714 41.2 0.002
Hs20556598 32.3 1.0
CE18785 32.0 1.4
Hs22061364 29.3 8.2
AtMi044 29.3 9.6
> 7292250
Length=256
Score = 382 bits (981), Expect = 5e-106, Method: Compositional matrix adjust.
Identities = 178/250 (71%), Positives = 216/250 (86%), Gaps = 0/250 (0%)
Query 14 MGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAV 73
MGRVIR+QRKG GSVF+AH KRKGAAKLR+LD+AER+GYI+G+V I HDPGRGAPLAV
Sbjct 1 MGRVIRAQRKGAGSVFKAHVKKRKGAAKLRSLDFAERSGYIRGVVKDIIHDPGRGAPLAV 60
Query 74 VSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKA 133
V FRD YRY+I KE +A EG+HTGQF++CG+KA L IGNV+PL ++PEGT++C++EEK
Sbjct 61 VHFRDPYRYKIRKELFIAPEGMHTGQFVYCGRKATLQIGNVMPLSQMPEGTIICNLEEKT 120
Query 134 GDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLL 193
GDRG +A+TSG +ATV+ H+++T KTRV+LPSGA+K +PS +R ++G+VAGGGRIDKP+L
Sbjct 121 GDRGRLARTSGNYATVIAHNQDTKKTRVKLPSGAKKVVPSANRAMVGIVAGGGRIDKPIL 180
Query 194 KAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPGQKVGLIAA 253
KAG AYHKYK KRN WPKVRGVAMNPVEHPHGGGNHQHIG STV R G+KVGLIAA
Sbjct 181 KAGRAYHKYKVKRNSWPKVRGVAMNPVEHPHGGGNHQHIGKASTVKRGTSAGRKVGLIAA 240
Query 254 RRTGLLRGGR 263
RRTG +RGG+
Sbjct 241 RRTGRIRGGK 250
> CE18478
Length=260
Score = 372 bits (955), Expect = 5e-103, Method: Compositional matrix adjust.
Identities = 174/258 (67%), Positives = 212/258 (82%), Gaps = 0/258 (0%)
Query 14 MGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAV 73
MGR IR QRKG G +F++H RKGA+KLR LDYAER+GYIKGLV I HDPGRGAPLA+
Sbjct 1 MGRRIRIQRKGAGGIFKSHNKHRKGASKLRPLDYAERHGYIKGLVKDIIHDPGRGAPLAI 60
Query 74 VSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKA 133
++FRD Y+Y+ K ++A EG+HTGQFIHCG KA + IGN++P+G +PEGT +C+VE K+
Sbjct 61 IAFRDPYKYKTVKTTVVAAEGMHTGQFIHCGAKAQIQIGNIVPVGTLPEGTTICNVENKS 120
Query 134 GDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLL 193
GDRG IA+ SG +ATV+ H+ +T KTR+RLPSGA+K + S +R +IGLVAGGGR DKPLL
Sbjct 121 GDRGVIARASGNYATVIAHNPDTKKTRIRLPSGAKKVVQSVNRAMIGLVAGGGRTDKPLL 180
Query 194 KAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPGQKVGLIAA 253
KAG +YHKYKAKRN WP+VRGVAMNPVEHPHGGGNHQHIGHPSTV R A G+KVGLIAA
Sbjct 181 KAGRSYHKYKAKRNSWPRVRGVAMNPVEHPHGGGNHQHIGHPSTVRRDASAGKKVGLIAA 240
Query 254 RRTGLLRGGRKTKLVGKD 271
RRTG +RGG+ K ++
Sbjct 241 RRTGRIRGGKPVKFTKEE 258
> At4g36130
Length=258
Score = 362 bits (929), Expect = 5e-100, Method: Compositional matrix adjust.
Identities = 167/248 (67%), Positives = 203/248 (81%), Gaps = 0/248 (0%)
Query 14 MGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAV 73
MGRVIR+QRKG GSVF++HTH RKG AK R+LD+ ERNGY+KG+VT I HDPGRGAPLA
Sbjct 1 MGRVIRAQRKGAGSVFKSHTHHRKGPAKFRSLDFGERNGYLKGVVTEIIHDPGRGAPLAR 60
Query 74 VSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKA 133
V+FR +R++ KE +A EG++TGQF++CGKKA L +GNVLPL +PEG V+C+VE
Sbjct 61 VAFRHPFRFKKQKELFVAAEGMYTGQFLYCGKKATLVVGNVLPLRSIPEGAVICNVEHHV 120
Query 134 GDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLL 193
GDRG A+ SG +A V+ H+ + +R++LPSG++K +PS R +IG VAGGGR +KP+L
Sbjct 121 GDRGVFARASGDYAIVIAHNPDNDTSRIKLPSGSKKIVPSGCRAMIGQVAGGGRTEKPML 180
Query 194 KAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPGQKVGLIAA 253
KAG AYHKY+ KRNCWPKVRGVAMNPVEHPHGGGNHQHIGH STV R APPG+KVGLIAA
Sbjct 181 KAGNAYHKYRVKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHASTVRRDAPPGKKVGLIAA 240
Query 254 RRTGLLRG 261
RRTG LRG
Sbjct 241 RRTGRLRG 248
> At2g18020
Length=258
Score = 362 bits (928), Expect = 6e-100, Method: Compositional matrix adjust.
Identities = 169/248 (68%), Positives = 204/248 (82%), Gaps = 0/248 (0%)
Query 14 MGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAV 73
MGRVIR+QRKG GSVF++HTH RKG AK R+LD+ ERNGY+KG+VT I HDPGRGAPLA
Sbjct 1 MGRVIRAQRKGAGSVFKSHTHHRKGPAKFRSLDFGERNGYLKGVVTEIIHDPGRGAPLAR 60
Query 74 VSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKA 133
V+FR +R++ KE +A EG++TGQF++CGKKA L +GNVLPL +PEG VVC+VE
Sbjct 61 VTFRHPFRFKKQKELFVAAEGMYTGQFLYCGKKATLVVGNVLPLRSIPEGAVVCNVEHHV 120
Query 134 GDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLL 193
GDRG +A+ SG +A V+ H+ ++ TR++LPSG++K +PS R +IG VAGGGR +KP+L
Sbjct 121 GDRGVLARASGDYAIVIAHNPDSDTTRIKLPSGSKKIVPSGCRAMIGQVAGGGRTEKPML 180
Query 194 KAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPGQKVGLIAA 253
KAG AYHKY+ KRN WPKVRGVAMNPVEHPHGGGNHQHIGH STV R APPGQKVGLIAA
Sbjct 181 KAGNAYHKYRVKRNSWPKVRGVAMNPVEHPHGGGNHQHIGHASTVRRDAPPGQKVGLIAA 240
Query 254 RRTGLLRG 261
RRTG LRG
Sbjct 241 RRTGRLRG 248
> Hs4506663
Length=257
Score = 356 bits (913), Expect = 3e-98, Method: Compositional matrix adjust.
Identities = 177/248 (71%), Positives = 209/248 (84%), Gaps = 0/248 (0%)
Query 14 MGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAV 73
MGRVIR QRKG GSVFRAH RKGAA+LRA+D+AER+GYIKG+V I HDPGRGAPLA
Sbjct 1 MGRVIRGQRKGAGSVFRAHVKHRKGAARLRAVDFAERHGYIKGIVKDIIHDPGRGAPLAK 60
Query 74 VSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKA 133
V FRD YR++ E +A EG+HTGQF++CGKKA L IGNVLP+G +PEGT+VC +EEK
Sbjct 61 VVFRDPYRFKKRTELFIAAEGIHTGQFVYCGKKAQLNIGNVLPVGTMPEGTIVCCLEEKP 120
Query 134 GDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLL 193
GDRG +A+ SG +ATV+ H+ ET KTRV+LPSG++K + S +R ++G+VAGGGRIDKP+L
Sbjct 121 GDRGKLARASGNYATVISHNPETKKTRVKLPSGSKKVISSANRAVVGVVAGGGRIDKPIL 180
Query 194 KAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPGQKVGLIAA 253
KAG AYHKYKAKRNCWP+VRGVAMNPVEHP GGGNHQHIG PST+ R AP G+KVGLIAA
Sbjct 181 KAGRAYHKYKAKRNCWPRVRGVAMNPVEHPFGGGNHQHIGKPSTIRRDAPAGRKVGLIAA 240
Query 254 RRTGLLRG 261
RRTG LRG
Sbjct 241 RRTGRLRG 248
> SPBC839.04
Length=253
Score = 347 bits (891), Expect = 1e-95, Method: Compositional matrix adjust.
Identities = 168/248 (67%), Positives = 211/248 (85%), Gaps = 1/248 (0%)
Query 14 MGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAV 73
MGRVIR+QRK G +F+AHT RKGAA+LR LD+AER+GYI+G+V I HDPGRGAPLA
Sbjct 1 MGRVIRAQRKS-GGIFQAHTRLRKGAAQLRTLDFAERHGYIRGVVQKIIHDPGRGAPLAK 59
Query 74 VSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKA 133
V+FR+ Y YR + E +A EG++TGQF++CGK A LT+GNVLP+G++PEGT++ +VEEKA
Sbjct 60 VAFRNPYHYRTDVETFVATEGMYTGQFVYCGKNAALTVGNVLPVGEMPEGTIISNVEEKA 119
Query 134 GDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLL 193
GDRG + ++SG + +VGH +TGKTRV+LPSGA+K +PS +R ++G+VAGGGRIDKPLL
Sbjct 120 GDRGALGRSSGNYVIIVGHDVDTGKTRVKLPSGAKKVVPSSARGVVGIVAGGGRIDKPLL 179
Query 194 KAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPGQKVGLIAA 253
KAG A+HKY+ KRNCWP+ RGVAMNPV+HPHGGGNHQH+GH +TV R + PGQKVGLIAA
Sbjct 180 KAGRAFHKYRVKRNCWPRTRGVAMNPVDHPHGGGNHQHVGHSTTVPRQSAPGQKVGLIAA 239
Query 254 RRTGLLRG 261
RRTGLLRG
Sbjct 240 RRTGLLRG 247
> SPBC2F12.07c
Length=253
Score = 347 bits (891), Expect = 1e-95, Method: Compositional matrix adjust.
Identities = 168/248 (67%), Positives = 211/248 (85%), Gaps = 1/248 (0%)
Query 14 MGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAV 73
MGRVIR+QRK G +F+AHT RKGAA+LR LD+AER+GYI+G+V I HDPGRGAPLA
Sbjct 1 MGRVIRAQRKS-GGIFQAHTRLRKGAAQLRTLDFAERHGYIRGVVQKIIHDPGRGAPLAK 59
Query 74 VSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKA 133
V+FR+ Y YR + E +A EG++TGQF++CGK A LT+GNVLP+G++PEGT++ +VEEKA
Sbjct 60 VAFRNPYHYRTDVETFVATEGMYTGQFVYCGKNAALTVGNVLPVGEMPEGTIISNVEEKA 119
Query 134 GDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLL 193
GDRG + ++SG + +VGH +TGKTRV+LPSGA+K +PS +R ++G+VAGGGRIDKPLL
Sbjct 120 GDRGALGRSSGNYVIIVGHDVDTGKTRVKLPSGAKKVVPSSARGVVGIVAGGGRIDKPLL 179
Query 194 KAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPGQKVGLIAA 253
KAG A+HKY+ KRNCWP+ RGVAMNPV+HPHGGGNHQH+GH +TV R + PGQKVGLIAA
Sbjct 180 KAGRAFHKYRVKRNCWPRTRGVAMNPVDHPHGGGNHQHVGHSTTVPRQSAPGQKVGLIAA 239
Query 254 RRTGLLRG 261
RRTGLLRG
Sbjct 240 RRTGLLRG 247
> YIL018w
Length=254
Score = 346 bits (887), Expect = 3e-95, Method: Compositional matrix adjust.
Identities = 172/253 (67%), Positives = 208/253 (82%), Gaps = 0/253 (0%)
Query 14 MGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAV 73
MGRVIR+QRKG GS+F +HT R+GAAKLR LDYAER+GYI+G+V I HD GRGAPLA
Sbjct 1 MGRVIRNQRKGAGSIFTSHTRLRQGAAKLRTLDYAERHGYIRGIVKQIVHDSGRGAPLAK 60
Query 74 VSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKA 133
V FRD Y+YR+ +E +A EG+HTGQFI+ GKKA L +GNVLPLG VPEGT+V +VEEK
Sbjct 61 VVFRDPYKYRLREEIFIANEGVHTGQFIYAGKKASLNVGNVLPLGSVPEGTIVSNVEEKP 120
Query 134 GDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLL 193
GDRG +A+ SG + ++GH+ + KTRVRLPSGA+K + S +R +IG++AGGGR+DKPLL
Sbjct 121 GDRGALARASGNYVIIIGHNPDENKTRVRLPSGAKKVISSDARGVIGVIAGGGRVDKPLL 180
Query 194 KAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPGQKVGLIAA 253
KAG A+HKY+ KRN WPK RGVAMNPV+HPHGGGNHQHIG ST+SR A GQK GLIAA
Sbjct 181 KAGRAFHKYRLKRNSWPKTRGVAMNPVDHPHGGGNHQHIGKASTISRGAVSGQKAGLIAA 240
Query 254 RRTGLLRGGRKTK 266
RRTGLLRG +KT+
Sbjct 241 RRTGLLRGSQKTQ 253
> YFR031c-a
Length=254
Score = 346 bits (887), Expect = 3e-95, Method: Compositional matrix adjust.
Identities = 172/253 (67%), Positives = 208/253 (82%), Gaps = 0/253 (0%)
Query 14 MGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAV 73
MGRVIR+QRKG GS+F +HT R+GAAKLR LDYAER+GYI+G+V I HD GRGAPLA
Sbjct 1 MGRVIRNQRKGAGSIFTSHTRLRQGAAKLRTLDYAERHGYIRGIVKQIVHDSGRGAPLAK 60
Query 74 VSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKA 133
V FRD Y+YR+ +E +A EG+HTGQFI+ GKKA L +GNVLPLG VPEGT+V +VEEK
Sbjct 61 VVFRDPYKYRLREEIFIANEGVHTGQFIYAGKKASLNVGNVLPLGSVPEGTIVSNVEEKP 120
Query 134 GDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLL 193
GDRG +A+ SG + ++GH+ + KTRVRLPSGA+K + S +R +IG++AGGGR+DKPLL
Sbjct 121 GDRGALARASGNYVIIIGHNPDENKTRVRLPSGAKKVISSDARGVIGVIAGGGRVDKPLL 180
Query 194 KAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPGQKVGLIAA 253
KAG A+HKY+ KRN WPK RGVAMNPV+HPHGGGNHQHIG ST+SR A GQK GLIAA
Sbjct 181 KAGRAFHKYRLKRNSWPKTRGVAMNPVDHPHGGGNHQHIGKASTISRGAVSGQKAGLIAA 240
Query 254 RRTGLLRGGRKTK 266
RRTGLLRG +KT+
Sbjct 241 RRTGLLRGSQKTQ 253
> At3g51190
Length=260
Score = 342 bits (876), Expect = 7e-94, Method: Compositional matrix adjust.
Identities = 162/239 (67%), Positives = 191/239 (79%), Gaps = 1/239 (0%)
Query 14 MGRVIRSQRKGR-GSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLA 72
MGRVIR+QRKG GSVF++HTH RKG AK R+LDY ERNGY+KGLVT I HDPGRGAPLA
Sbjct 1 MGRVIRAQRKGAAGSVFKSHTHHRKGPAKFRSLDYGERNGYLKGLVTEIIHDPGRGAPLA 60
Query 73 VVSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEK 132
V+FR +RY KE +A EG++TGQ+++CGKKA+L +GNVLPLG +PEG V+C+VE
Sbjct 61 RVAFRHPFRYMKQKELFVAAEGMYTGQYLYCGKKANLMVGNVLPLGSIPEGAVICNVELH 120
Query 133 AGDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPL 192
GDRG +A+ SG +A V+ H+ E+ TRV+LPSG++K LPS R +IG VAGGGR +KP
Sbjct 121 VGDRGALARASGDYAIVIAHNPESNTTRVKLPSGSKKILPSACRAMIGQVAGGGRTEKPF 180
Query 193 LKAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPGQKVGLI 251
LKAG AYHKYKAKRNCWP VRGVAMNPVEHPHGGGNHQHIGH STV R G KVG I
Sbjct 181 LKAGNAYHKYKAKRNCWPVVRGVAMNPVEHPHGGGNHQHIGHASTVRRDKSAGAKVGQI 239
> Hs17444499
Length=239
Score = 225 bits (573), Expect = 9e-59, Method: Compositional matrix adjust.
Identities = 128/253 (50%), Positives = 161/253 (63%), Gaps = 18/253 (7%)
Query 14 MGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAV 73
MG ++ QRKG G+ F A A L + +GY K V H PG GAPLA
Sbjct 1 MGLMVCGQRKGAGAAFPAQR-----ACAL----WTAPSGY-KSTVKGAIHYPGCGAPLAK 50
Query 74 VSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKA 133
V FR+A ++ + +A EG+H GQF++CGKK L IG+VL + +PE T VC ++EK
Sbjct 51 VVFRNAPKW---TQLFIAAEGIHMGQFVYCGKKVQLNIGSVLSVDTMPESTTVCCLKEKP 107
Query 134 GDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLL 193
DRG +A+ SG ATV+ H+ ET KT A +R + G+VA GGR DK +L
Sbjct 108 EDRGKLARASGNGATVISHNPETQKTHE-----AALLRQQANRAVGGVVARGGRNDKAIL 162
Query 194 KAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPGQKVGLIAA 253
KAG A+HKYKAKRNCWP+VRGVAMN VEHP GGGNHQHIG PST+ R AP +KVGLIA
Sbjct 163 KAGRAHHKYKAKRNCWPRVRGVAMNLVEHPFGGGNHQHIGKPSTIRRDAPADRKVGLIAV 222
Query 254 RRTGLLRGGRKTK 266
R+TG LRG + +
Sbjct 223 RQTGRLRGAKTVQ 235
> ECU01g0310
Length=239
Score = 200 bits (508), Expect = 3e-51, Method: Compositional matrix adjust.
Identities = 114/252 (45%), Positives = 158/252 (62%), Gaps = 15/252 (5%)
Query 14 MGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAV 73
MG++ R R+ A +R+ K+ L Y +G V I H+ RGAP+A+
Sbjct 1 MGKITRRTRE-------AKKKQRQPIVKV-CLSYPIIPQVEEGEVIDIVHERSRGAPVAI 52
Query 74 VSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKA 133
+SF++ + + A EG++TGQ I G A + IGN+ + VPEG V SVE
Sbjct 53 ISFKE------DDCMVPASEGMYTGQKILIGDDAPIDIGNITKIKNVPEGMAVNSVESVY 106
Query 134 GDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLL 193
GD GT A +G+++ VV H +ET +T +++PSG + T+ S RCI+G+VAGGG DKPLL
Sbjct 107 GDGGTFAMVNGSYSLVVNHRKETNETVIKIPSGKKLTVSSECRCIVGVVAGGGIHDKPLL 166
Query 194 KAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVS-RTAPPGQKVGLIA 252
KA A++K KA+ + +P+VRGVAMNPVEH HGGGNHQH+G P+TVS + QK+GL+
Sbjct 167 KASVAHYKAKARGHVFPRVRGVAMNPVEHIHGGGNHQHVGKPTTVSKKDISQEQKIGLVG 226
Query 253 ARRTGLLRGGRK 264
ARRTG G RK
Sbjct 227 ARRTGYRVGSRK 238
> Hs17459950
Length=124
Score = 123 bits (308), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 68/121 (56%), Positives = 89/121 (73%), Gaps = 9/121 (7%)
Query 126 VCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGG 185
+C +EEK D G + + SG +ATV+ H+ ET K+RV+LPSG++K S +R ++G+VAGG
Sbjct 1 MCCLEEKPRDHGKLTQASGNYATVISHNPETKKSRVKLPSGSKKVTSSANRAVVGVVAGG 60
Query 186 GRIDKPLLKAGTAYHKYKAKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRTAPPG 245
G+IDKP+LKAG AYH+YKAKRNCWP+VRG GGG+HQHIG PST+ R A G
Sbjct 61 GQIDKPILKAGWAYHRYKAKRNCWPQVRGG---------GGGSHQHIGKPSTICRDASAG 111
Query 246 Q 246
Q
Sbjct 112 Q 112
> AtCh087
Length=274
Score = 92.8 bits (229), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 60/189 (31%), Positives = 92/189 (48%), Gaps = 20/189 (10%)
Query 43 RALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLAVEGLHTGQFIH 102
R +D+ I G + +IE+DP R A + ++ + D K +L G G I
Sbjct 61 RKIDFRRNAKDIYGRIVTIEYDPNRNAYICLIHYGDG-----EKRYILHPRGAIIGDTIV 115
Query 103 CGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVR 162
G + + +GN LPL +P GT + ++E G G +A+ +G A ++ ++E ++
Sbjct 116 SGTEVPIKMGNALPLTDMPLGTAIHNIEITLGRGGQLARAAGAVAKLI--AKEGKSATLK 173
Query 163 LPSGARKTLPSRSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW----PKVRGVAMN 218
LPSG + + +G V G K L +AG+ CW P VRGV MN
Sbjct 174 LPSGEVRLISKNCSATVGQVGNVGVNQKSLGRAGS---------KCWLGKRPVVRGVVMN 224
Query 219 PVEHPHGGG 227
PV+HPHGGG
Sbjct 225 PVDHPHGGG 233
> AtCh063
Length=274
Score = 92.8 bits (229), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 60/189 (31%), Positives = 92/189 (48%), Gaps = 20/189 (10%)
Query 43 RALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLAVEGLHTGQFIH 102
R +D+ I G + +IE+DP R A + ++ + D K +L G G I
Sbjct 61 RKIDFRRNAKDIYGRIVTIEYDPNRNAYICLIHYGDG-----EKRYILHPRGAIIGDTIV 115
Query 103 CGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVR 162
G + + +GN LPL +P GT + ++E G G +A+ +G A ++ ++E ++
Sbjct 116 SGTEVPIKMGNALPLTDMPLGTAIHNIEITLGRGGQLARAAGAVAKLI--AKEGKSATLK 173
Query 163 LPSGARKTLPSRSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW----PKVRGVAMN 218
LPSG + + +G V G K L +AG+ CW P VRGV MN
Sbjct 174 LPSGEVRLISKNCSATVGQVGNVGVNQKSLGRAGS---------KCWLGKRPVVRGVVMN 224
Query 219 PVEHPHGGG 227
PV+HPHGGG
Sbjct 225 PVDHPHGGG 233
> YEL050c
Length=393
Score = 72.4 bits (176), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 62/210 (29%), Positives = 93/210 (44%), Gaps = 39/210 (18%)
Query 43 RALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLAVEGLHTGQFIH 102
R +D+ G + V IE+DPGR + +A++ ++A +GL G +
Sbjct 153 RLIDFNRWEGGAQ-TVQRIEYDPGRSSHIALLKHNTTGEL----SYIIACDGLRPGDVVE 207
Query 103 -------------CGKKADLTI--------GNVLPLGKVPEGTVVCSVEEKAGDRGTIAK 141
G K D I GN LP+ +P GT++ +V G +
Sbjct 208 SFRRGIPQTLLNEMGGKVDPAILSVKTTQRGNCLPISMIPIGTIIHNVGITPVGPGKFCR 267
Query 142 TSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLLKAGTAYHK 201
++GT+A V+ E K VRL SG + + + IG+V+ ++ L KAG
Sbjct 268 SAGTYARVLAKLPEKKKAIVRLQSGEHRYVSLEAVATIGVVSNIDHQNRSLGKAG----- 322
Query 202 YKAKRNCW----PKVRGVAMNPVEHPHGGG 227
R+ W P VRGVAMN +HPHGGG
Sbjct 323 ----RSRWLGIRPTVRGVAMNKCDHPHGGG 348
> SPAC1F7.13c
Length=47
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 33/41 (80%), Positives = 37/41 (90%), Gaps = 0/41 (0%)
Query 221 EHPHGGGNHQHIGHPSTVSRTAPPGQKVGLIAARRTGLLRG 261
+HPHGGGNHQH+GH +TV R + PGQKVGLIAARRTGLLRG
Sbjct 1 DHPHGGGNHQHVGHSTTVPRQSAPGQKVGLIAARRTGLLRG 41
> SPCC16C4.15
Length=318
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 62/233 (26%), Positives = 102/233 (43%), Gaps = 50/233 (21%)
Query 41 KLRALDYAERNGYIKGL--VTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLAVEGLHTG 98
++R +D+ + + G+ V IE+DPGR +A+V ++ NK +LA +GL G
Sbjct 69 RIRLVDFERK---VPGVHRVIRIEYDPGRSGHIALVEKLNSET--ANKSYILACDGLREG 123
Query 99 QFIHCGKKA----------------------------DLTIGNVLPLGKVPEGTVVCSVE 130
+ + +L GN PL +P GTV+ ++
Sbjct 124 DTVESFRSLLKTGSNGEPVEMDPVLAAEIEDGTFAAKNLKPGNCFPLRLIPIGTVIHAIG 183
Query 131 EKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDK 190
+ + +++G+ A ++ + VRL SG + + S IG+V+ +
Sbjct 184 VNPNQKAKLCRSAGSSARIIAFDGKYAI--VRLQSGEERKILDTSFATIGVVSNIYWQHR 241
Query 191 PLLKAGTAYHKYKAKRNCW----PKVRGVAMNPVEHPHGGGNHQHIGHPSTVS 239
L KAG R+ W P VRG AMNP +HPHGGG + IG+ + S
Sbjct 242 QLGKAG---------RSRWLGIRPTVRGTAMNPCDHPHGGGGGKSIGNKPSQS 285
> At4g14250_2
Length=67
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/78 (41%), Positives = 43/78 (55%), Gaps = 12/78 (15%)
Query 150 VGHSEETGKTRVRLPSGARKTLPSRSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 209
+ +S + + LP ++KT+ S R +IG +A G K ++K RN W
Sbjct 1 IANSMISVTWDINLPLDSKKTVLSGCRVMIGQIASSGLTKKLMIK-----------RNMW 49
Query 210 PKVRGVAM-NPVEHPHGG 226
KVRGVAM NPVEHPHGG
Sbjct 50 AKVRGVAMMNPVEHPHGG 67
> 7294756
Length=294
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 36/115 (31%), Positives = 56/115 (48%), Gaps = 3/115 (2%)
Query 51 NGYIKGLVTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLT 110
G + LV + D R A +A+V+ D +Y + E M A + L T +FI
Sbjct 101 EGAQEELVLEVLRDGCRTAKVALVAVGDELKYILATENMKAGDILKTSRFIP-RIPVRPN 159
Query 111 IGNVLPLGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPS 165
G+ PLG +P GT + +E+ G + +GTF T++ +E K V+LPS
Sbjct 160 EGDAYPLGALPVGTRIHCLEKNPGQMCHLIHAAGTFGTILRKFDE--KVVVQLPS 212
> CE18483_2
Length=72
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 19/37 (51%), Positives = 28/37 (75%), Gaps = 0/37 (0%)
Query 144 GTFATVVGHSEETGKTRVRLPSGARKTLPSRSRCIIG 180
G +A V+ H+ +T KTR+RLPSGA+K + S +R +IG
Sbjct 1 GNYANVIAHNADTKKTRIRLPSGAKKVVQSVNRAMIG 37
> Hs21265070
Length=305
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 35/135 (25%), Positives = 62/135 (45%), Gaps = 5/135 (3%)
Query 32 HTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLA 91
H + + LR ++G + V + +DP R A +A+V+ R+ I E M A
Sbjct 100 HKQRYRMIDFLRFRPEETKSGPFEEKVIQVRYDPCRSADIALVAGGSRKRWIIATENMQA 159
Query 92 VEGLHTGQFIHCGKKADLT-IGNVLPLGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVV 150
+ + H G+ A G+ PLG +P GT++ +VE + G + +GT ++
Sbjct 160 GDTILNSN--HIGRMAVAAREGDAHPLGALPVGTLINNVESEPGRGAQYIRAAGTCGVLL 217
Query 151 GHSEETGKTRVRLPS 165
+ G ++LPS
Sbjct 218 --RKVNGTAIIQLPS 230
> HsM7705714
Length=353
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 35/135 (25%), Positives = 63/135 (46%), Gaps = 5/135 (3%)
Query 32 HTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLA 91
H + + LR ++G + V + +DP R A +A+V+ R+ I E M A
Sbjct 100 HKQRYRMIDFLRFRPEETKSGPFEEKVIQVRYDPCRSADIALVAGGSRKRWIIATENMQA 159
Query 92 VEGLHTGQFIHCGKKADLT-IGNVLPLGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVV 150
+ + H G+ A G+ PLG +P GT++ +VE + G + +GT ++
Sbjct 160 GDTILNSN--HIGRMAVAAREGDAHPLGALPVGTLINNVESEPGRGAQYIRAAGTCGVLL 217
Query 151 GHSEETGKTRVRLPS 165
+ G + ++LPS
Sbjct 218 --RKVNGHSIIQLPS 230
> Hs20556598
Length=427
Score = 32.3 bits (72), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 16/20 (80%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 242 APPGQKVGLIAARRTGLLRG 261
AP G+KVGLIA RRTG LRG
Sbjct 399 APAGRKVGLIAPRRTGRLRG 418
> CE18785
Length=561
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 5 NSSSSSSRKMGRVIRSQRKGRGSVFRAHTHKRKGAAKLRALDYAER 50
N+S ++GR RS +KG F HT+ K L+ LD A++
Sbjct 455 NNSEDYVHRIGRTGRSDKKGTAYTFFTHTNASKAKDLLKVLDEAKQ 500
> Hs22061364
Length=197
Score = 29.3 bits (64), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 31/54 (57%), Gaps = 2/54 (3%)
Query 112 GNVLPLGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPS 165
G+ PLG +P GT++ ++E + G + +GT + ++ + G T ++LPS
Sbjct 84 GDAHPLGALPVGTLINNMEIEPGRGAQYIQAAGTCSVLLW--KVNGTTVIQLPS 135
> AtMi044
Length=349
Score = 29.3 bits (64), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Query 24 GRGSVFRAHTHKRKGAAK--LRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 76
GR S R R G +K LR +D +R+ G+V SIE+DP R + +A V +
Sbjct 21 GRNSSGRITVFHRGGGSKRLLRRIDL-KRSTSSMGIVESIEYDPNRSSQIAPVRW 74
Lambda K H
0.317 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5843977590
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40