bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
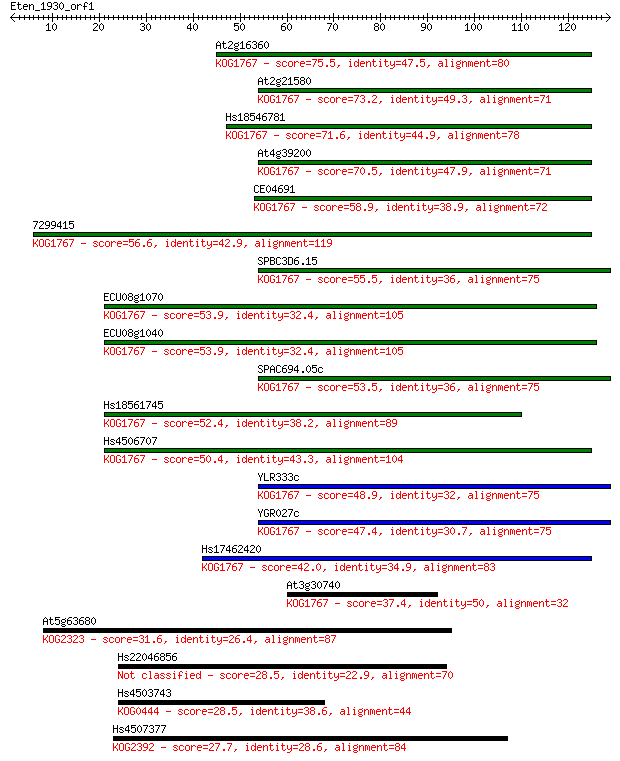
Query= Eten_1930_orf1
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
At2g16360 75.5 2e-14
At2g21580 73.2 1e-13
Hs18546781 71.6 3e-13
At4g39200 70.5 8e-13
CE04691 58.9 2e-09
7299415 56.6 1e-08
SPBC3D6.15 55.5 2e-08
ECU08g1070 53.9 7e-08
ECU08g1040 53.9 7e-08
SPAC694.05c 53.5 9e-08
Hs18561745 52.4 2e-07
Hs4506707 50.4 8e-07
YLR333c 48.9 2e-06
YGR027c 47.4 6e-06
Hs17462420 42.0 3e-04
At3g30740 37.4 0.007
At5g63680 31.6 0.35
Hs22046856 28.5 2.9
Hs4503743 28.5 3.0
Hs4507377 27.7 5.6
> At2g16360
Length=125
Score = 75.5 bits (184), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/80 (47%), Positives = 52/80 (65%), Gaps = 1/80 (1%)
Query 45 KKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELY 104
+K SKGK K+K+N V+FD T +K +SE PK KL+TP + +R RIN SLAR+ IR+L
Sbjct 42 RKRSKGKQKEKVNNMVLFDQATYDKLMSEAPKFKLITPSILSDRLRINGSLARKAIRDLM 101
Query 105 RQQQLRPLGDQHHSHYIFTR 124
+ +R + H S I TR
Sbjct 102 VKGTIR-MVSTHSSQQINTR 120
> At2g21580
Length=108
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/71 (49%), Positives = 47/71 (66%), Gaps = 1/71 (1%)
Query 54 DKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLG 113
+K+N V+FD GT +K L+E PK KL+TP + +R RIN SLARR IREL + +R +
Sbjct 35 EKVNNMVLFDQGTYDKLLTEAPKFKLITPSILSDRMRINGSLARRAIRELMAKGLIR-MV 93
Query 114 DQHHSHYIFTR 124
H S I+TR
Sbjct 94 SAHSSQQIYTR 104
> Hs18546781
Length=145
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 35/78 (44%), Positives = 48/78 (61%), Gaps = 1/78 (1%)
Query 47 WSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQ 106
WS+GK DKLN V+FD T K EVPK KL+TP + ER +I SLAR ++EL +
Sbjct 55 WSRGKVWDKLNSLVLFDKATYNKLCKEVPKYKLITPAVVSERLKIRGSLARAALQELLSK 114
Query 107 QQLRPLGDQHHSHYIFTR 124
++ L +H + I+TR
Sbjct 115 GLIK-LVSKHRAQVIYTR 131
> At4g39200
Length=108
Score = 70.5 bits (171), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 34/71 (47%), Positives = 46/71 (64%), Gaps = 1/71 (1%)
Query 54 DKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLG 113
+K+N V+FD T +K L+E PK KL+TP + +R RIN SLARR IREL + +R +
Sbjct 35 EKVNNMVLFDQATYDKLLTEAPKFKLITPSILSDRMRINGSLARRAIRELMAKGVIRMVA 94
Query 114 DQHHSHYIFTR 124
H S I+TR
Sbjct 95 -AHSSQQIYTR 104
> CE04691
Length=117
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 44/72 (61%), Gaps = 1/72 (1%)
Query 53 KDKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPL 112
+DKLN V+FD T +K EV KL+TP + ER ++ ASLA+ G++EL + ++ +
Sbjct 37 RDKLNNMVLFDQATYDKLYKEVITYKLITPSVVSERLKVRASLAKAGLKELQAKGLVKCV 96
Query 113 GDQHHSHYIFTR 124
HH ++TR
Sbjct 97 V-HHHGQVVYTR 107
> 7299415
Length=136
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 51/119 (42%), Positives = 66/119 (55%), Gaps = 3/119 (2%)
Query 6 FYSFLFNSVFKNCDRMAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAG 65
F+ F K+ A ++ K Q + G K KKKWSKGK +DKLN V+FD
Sbjct 14 FFFATFQPPKKDAKSSA--KQPQKTQKKKEGSGGGKAKKKKWSKGKVRDKLNNQVLFDKA 71
Query 66 TLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
T EK EVP KL+TP + ER +I SLA+R + EL R++ L QHHS I+TR
Sbjct 72 TYEKLYKEVPAYKLITPSVVSERLKIRGSLAKRALIEL-REKGLIKQVVQHHSQVIYTR 129
> SPBC3D6.15
Length=88
Score = 55.5 bits (132), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 44/75 (58%), Gaps = 1/75 (1%)
Query 54 DKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLG 113
DK +A +FD +++ EVP K ++ + +R +IN SLAR IR+L + ++ +
Sbjct 14 DKAQHATVFDKSIIDRINKEVPAFKFISVSVLVDRMKINGSLARIAIRDLAERGVIQKV- 72
Query 114 DQHHSHYIFTRPAAA 128
DQH I+TR AA+
Sbjct 73 DQHSKQAIYTRAAAS 87
> ECU08g1070
Length=109
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 34/108 (31%), Positives = 58/108 (53%), Gaps = 5/108 (4%)
Query 21 MAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLV 80
M K +++KE+ A AA+G++K KKKW G+ K+++ AV L K +V + +V
Sbjct 1 MVKKIQESKEKKALKAASGTRKDKKKWGDGRKKEEVRRAVTVSEELLAKVRKDVGRASVV 60
Query 81 TPFSICERFRINASLARRGIRELYRQ---QQLRPLGDQHHSHYIFTRP 125
T + I R+ +N +A +R L + QQ+ LG++ + Y R
Sbjct 61 TRYMIGSRYNLNLGVAENVLRHLSNEGVVQQV--LGNRRMTIYAGCRA 106
> ECU08g1040
Length=109
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 34/108 (31%), Positives = 58/108 (53%), Gaps = 5/108 (4%)
Query 21 MAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLV 80
M K +++KE+ A AA+G++K KKKW G+ K+++ AV L K +V + +V
Sbjct 1 MVKKIQESKEKKALKAASGTRKDKKKWGDGRKKEEVRRAVTVSEELLAKVRKDVGRASVV 60
Query 81 TPFSICERFRINASLARRGIRELYRQ---QQLRPLGDQHHSHYIFTRP 125
T + I R+ +N +A +R L + QQ+ LG++ + Y R
Sbjct 61 TRYMIGSRYNLNLGVAENVLRHLSNEGVVQQV--LGNRRMTIYAGCRA 106
> SPAC694.05c
Length=89
Score = 53.5 bits (127), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 43/75 (57%), Gaps = 1/75 (1%)
Query 54 DKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLG 113
DK +A +FD +++ EVP K ++ + +R +IN SLAR IR+L + ++ +
Sbjct 14 DKAQHATVFDKSIIDRINKEVPAFKFISVSVLVDRMKINGSLARIAIRDLAERGVIQKV- 72
Query 114 DQHHSHYIFTRPAAA 128
DQH I+TR AA
Sbjct 73 DQHSKQAIYTRVPAA 87
> Hs18561745
Length=113
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/101 (33%), Positives = 49/101 (48%), Gaps = 17/101 (16%)
Query 21 MAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKS------------KDKLNYAVMFDAGTLE 68
M PK+ + K+ A +KK K +K +DKLN V+FD T +
Sbjct 1 MPPKDDEKKD-----AGKSAKKDKDPVNKSGGKAKKKKWSKGKVRDKLNNLVLFDKATYD 55
Query 69 KFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQL 109
K EVP KL+TP + ER +I SLAR ++ +R Q +
Sbjct 56 KLCKEVPNYKLITPAVVSERLKIRGSLARAALQAKHRAQVI 96
> Hs4506707
Length=125
Score = 50.4 bits (119), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 45/112 (40%), Positives = 62/112 (55%), Gaps = 9/112 (8%)
Query 21 MAPKEKKTKEQIAAAAAAG--------SKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLS 72
M PK+ K K+ +A K KKKWSKGK +DKLN V+FD T +K
Sbjct 1 MPPKDDKKKKDAGKSAKKDKDPVNKSGGKAKKKKWSKGKVRDKLNNLVLFDKATYDKLCK 60
Query 73 EVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
EVP KL+TP + ER +I SLAR ++EL + ++ L +H + I+TR
Sbjct 61 EVPNYKLITPAVVSERLKIRGSLARAALQELLSKGLIK-LVSKHRAQVIYTR 111
> YLR333c
Length=108
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 43/75 (57%), Gaps = 1/75 (1%)
Query 54 DKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLG 113
D+ +AV+ D ++ L EVP + V+ + +R +I SLAR +R L ++ ++P+
Sbjct 34 DRAQHAVILDQEKYDRILKEVPTYRYVSVSVLVDRLKIGGSLARIALRHLEKEGIIKPIS 93
Query 114 DQHHSHYIFTRPAAA 128
+H I+TR AA+
Sbjct 94 -KHSKQAIYTRAAAS 107
> YGR027c
Length=108
Score = 47.4 bits (111), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 42/75 (56%), Gaps = 1/75 (1%)
Query 54 DKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLG 113
D+ +AV+ D ++ L EVP + V+ + +R +I SLAR +R L ++ ++P+
Sbjct 34 DRAQHAVILDQEKYDRILKEVPTYRYVSVSVLVDRLKIGGSLARIALRHLEKEGIIKPIS 93
Query 114 DQHHSHYIFTRPAAA 128
+H I+TR A+
Sbjct 94 -KHSKQAIYTRATAS 107
> Hs17462420
Length=102
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/83 (34%), Positives = 40/83 (48%), Gaps = 22/83 (26%)
Query 42 KSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIR 101
K+KK+WSKGK DKLN V+FD T R I SLAR ++
Sbjct 28 KAKKEWSKGKIWDKLNNQVLFDNAT---------------------RLEIRGSLARAALQ 66
Query 102 ELYRQQQLRPLGDQHHSHYIFTR 124
EL + ++ L +H + I+TR
Sbjct 67 ELLGKGLIK-LVSKHRARVIYTR 88
> At3g30740
Length=53
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 60 VMFDAGTLEKFLSEVPKMKLVTPFSICERFRI 91
V+FD T EK L E PK KL+TP + +R R+
Sbjct 2 VLFDQATYEKLLIEAPKFKLITPSILSDRMRV 33
> At5g63680
Length=510
Score = 31.6 bits (70), Expect = 0.35, Method: Composition-based stats.
Identities = 23/91 (25%), Positives = 40/91 (43%), Gaps = 14/91 (15%)
Query 8 SFLFNSVFKNCDRMAPKEKKTKEQIAAAAAAGSKKSKKK----WSKGKSKDKLNYAVMFD 63
S +N++FK R P T E +A++A + K+K K ++G + KL
Sbjct 358 SLDYNTIFKEMIRATPLPMSTLESLASSAVRTANKAKAKLIIVLTRGGTTAKL------- 410
Query 64 AGTLEKFLSEVPKMKLVTPFSICERFRINAS 94
+ K+ VP + +V P + F + S
Sbjct 411 ---VAKYRPAVPILSVVVPVFTSDTFNWSCS 438
> Hs22046856
Length=197
Score = 28.5 bits (62), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 16/70 (22%), Positives = 31/70 (44%), Gaps = 0/70 (0%)
Query 24 KEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLVTPF 83
+E+ ++ +A A + S+++W LN+ ++ G L L K K T F
Sbjct 63 RERTARDGVAGLAREVLQSSREEWKHHTPSRHLNFLLLMTTGALSSLLDSDGKEKEHTIF 122
Query 84 SICERFRINA 93
+ RI++
Sbjct 123 ELLIGSRISS 132
> Hs4503743
Length=1269
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query 24 KEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMF--DAGTL 67
+EK K++ +A A A S K ++ W +G K +L+Y+ F D G L
Sbjct 454 QEKNKKQEESADARAPSGKVRR-WDQGLEKPRLDYSEFFTEDVGQL 498
> Hs4507377
Length=415
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 24/84 (28%), Positives = 38/84 (45%), Gaps = 24/84 (28%)
Query 23 PKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLVTP 82
PKE Q+ + AA S K+ KKW++ + G ++ F VPK
Sbjct 272 PKEG----QMESVEAAMSSKTLKKWNR-----------LLQKGWVDLF---VPK------ 307
Query 83 FSICERFRINASLARRGIRELYRQ 106
FSI + + A+L + GI+ Y +
Sbjct 308 FSISATYDLGATLLKMGIQHAYSE 331
Lambda K H
0.322 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1213511838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40