bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1915_orf2
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
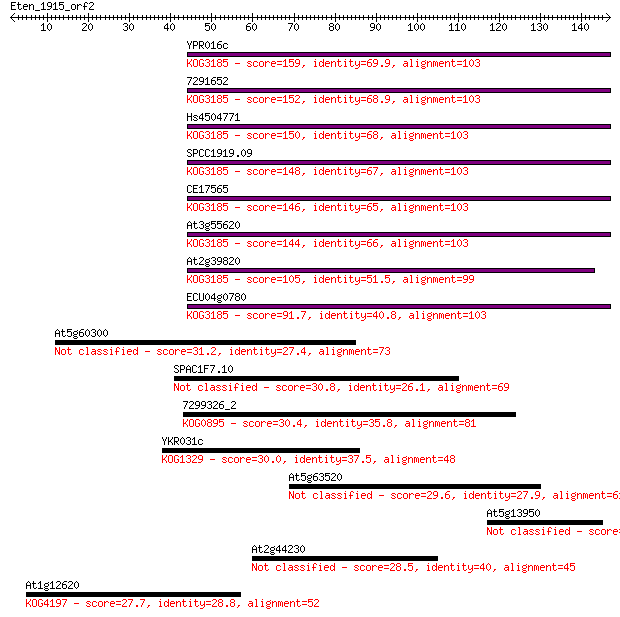
YPR016c 159 2e-39
7291652 152 2e-37
Hs4504771 150 1e-36
SPCC1919.09 148 3e-36
CE17565 146 1e-35
At3g55620 144 7e-35
At2g39820 105 3e-23
ECU04g0780 91.7 4e-19
At5g60300 31.2 0.74
SPAC1F7.10 30.8 0.87
7299326_2 30.4 1.2
YKR031c 30.0 1.7
At5g63520 29.6 2.3
At5g13950 28.9 4.0
At2g44230 28.5 5.1
At1g12620 27.7 7.3
> YPR016c
Length=245
Score = 159 bits (401), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 72/103 (69%), Positives = 86/103 (83%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA RT FE+SNE+GVF KLTN+YCLVA GGSE+FYSA EAELG IP+VH ++AGT +IG
Sbjct 1 MATRTQFENSNEIGVFSKLTNTYCLVAVGGSENFYSAFEAELGDAIPIVHTTIAGTRIIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+ GN+ GLLVP+ T+D ELQ LRNSLPD V ++RVEERLSA
Sbjct 61 RMTAGNRRGLLVPTQTTDQELQHLRNSLPDSVKIQRVEERLSA 103
> 7291652
Length=245
Score = 152 bits (385), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 71/103 (68%), Positives = 86/103 (83%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA+R FE+++++GVF KLTN+YCLVA GGSE FYSA EAELG IPVVHA+V G +IG
Sbjct 1 MALRVQFENNDDIGVFTKLTNTYCLVAIGGSETFYSAFEAELGDTIPVVHANVGGCRIIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+ VGN++GLLVP+ST+D ELQ LRNSLPD V + RVEERLSA
Sbjct 61 RLTVGNRNGLLVPNSTTDEELQHLRNSLPDAVKIYRVEERLSA 103
> Hs4504771
Length=245
Score = 150 bits (378), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 70/103 (67%), Positives = 85/103 (82%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA+R FE++ E+G F KLTN+YCLVA GGSE+FYS E EL IPVVHAS+AG +IG
Sbjct 1 MAVRASFENNCEIGCFAKLTNTYCLVAIGGSENFYSVFEGELSDTIPVVHASIAGCRIIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+CVGN+HGLLVP++T+D ELQ +RNSLPD V +RRVEERLSA
Sbjct 61 RMCVGNRHGLLVPNNTTDQELQHIRNSLPDTVQIRRVEERLSA 103
> SPCC1919.09
Length=244
Score = 148 bits (374), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 69/103 (66%), Positives = 83/103 (80%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA+R FE+SNE+GVF LTNSY LVA GGSE+FYS EAELG +PVVH ++ GT +IG
Sbjct 1 MALRAQFENSNEIGVFSNLTNSYALVALGGSENFYSVFEAELGDVVPVVHTTIGGTRIIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+ GN+ GLLVPSST+D ELQ LRNSLPD V ++RV+ERLSA
Sbjct 61 RLTCGNRKGLLVPSSTTDNELQHLRNSLPDPVKIQRVDERLSA 103
> CE17565
Length=246
Score = 146 bits (369), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 67/103 (65%), Positives = 85/103 (82%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA+R +E SN+VGVF LTNSYCLV GG+++FYS +EAEL IPVVH S+A T ++G
Sbjct 1 MALRVDYEGSNDVGVFCTLTNSYCLVGVGGTQNFYSILEAELSDLIPVVHTSIASTRIVG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+ VGN+HGLLVP++T+D ELQ LRNSLPD V++RRV+ERLSA
Sbjct 61 RLTVGNRHGLLVPNATTDQELQHLRNSLPDEVAIRRVDERLSA 103
> At3g55620
Length=245
Score = 144 bits (362), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 68/103 (66%), Positives = 83/103 (80%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA R FE++ EVGVF KLTN+YCLVA GGSE+FYSA E+EL IP+V S+ GT +IG
Sbjct 1 MATRLQFENNCEVGVFSKLTNAYCLVAIGGSENFYSAFESELADVIPIVKTSIGGTRIIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+C GNK+GLLVP +T+D ELQ LRNSLPD V V+R++ERLSA
Sbjct 61 RLCAGNKNGLLVPHTTTDQELQHLRNSLPDQVVVQRIDERLSA 103
> At2g39820
Length=247
Score = 105 bits (263), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 51/101 (50%), Positives = 74/101 (73%), Gaps = 2/101 (1%)
Query 44 MAIRTHFESSN-EVGVFGKLTNSYCLV-ASGGSEHFYSAIEAELGPHIPVVHASVAGTAV 101
MA R ++++N E+GVF KLTN+YCLV A+ S +F++ E++L IP+V S+ G+
Sbjct 1 MATRLQYDNNNCEIGVFSKLTNAYCLVSATSASANFFTGYESKLKGVIPIVTTSIGGSGT 60
Query 102 IGRVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEE 142
IG +CVGNK+GLL+ + +D ELQ LR+SLPD V V+R+EE
Sbjct 61 IGSLCVGNKNGLLLSHTITDQELQHLRDSLPDEVVVQRIEE 101
> ECU04g0780
Length=248
Score = 91.7 bits (226), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 42/103 (40%), Positives = 65/103 (63%), Gaps = 2/103 (1%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
M+ R FE S+E+G + LTN+YC++ S + ++ + IP+V ++ +G
Sbjct 8 MSYRIDFEGSSEIGAYMSLTNTYCVIGRSQSNNVLKFLQENVA--IPIVETTINSIRSVG 65
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
C GN+HGLLVP + +D E+ +RNSLP+ V VRR+EERL+A
Sbjct 66 SQCRGNRHGLLVPHTITDQEIMHIRNSLPEDVVVRRIEERLNA 108
> At5g60300
Length=718
Score = 31.2 bits (69), Expect = 0.74, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 36/76 (47%), Gaps = 10/76 (13%)
Query 12 FLRSATVTFGPIFLCGIIFRPGWAAFSSLVNKMAIR---THFESSNEVGVFGKLTNSYCL 68
F S ++F F+C ++ +PG+ +V ++ TH ES+ +G+F TN
Sbjct 73 FSSSGPLSFSTHFVCALVPKPGFEGGHGIVFVLSPSMDFTHAESTRYLGIFNASTN---- 128
Query 69 VASGGSEHFYSAIEAE 84
G S + A+E +
Sbjct 129 ---GSSSYHVLAVELD 141
> SPAC1F7.10
Length=238
Score = 30.8 bits (68), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 32/69 (46%), Gaps = 6/69 (8%)
Query 41 VNKMAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTA 100
V+ + RT + NE+G + CL G H +E +GP+IP++ + AG
Sbjct 161 VDALLARTALRAVNEMG-----ADVICL-GCAGMTHMAHVLEKAVGPNIPIIDGTKAGVE 214
Query 101 VIGRVCVGN 109
++ + N
Sbjct 215 LLASLVRMN 223
> 7299326_2
Length=4574
Score = 30.4 bits (67), Expect = 1.2, Method: Composition-based stats.
Identities = 29/89 (32%), Positives = 39/89 (43%), Gaps = 17/89 (19%)
Query 43 KMAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYS--------AIEAELGPHIPVVHA 94
++ I HF SS E K S C+V + G+ YS IE E G H
Sbjct 467 QLCILPHFNSSPE----HKDAGSICVVCADGTVEIYSLADFQGTMVIEEE-GEHF----E 517
Query 95 SVAGTAVIGRVCVGNKHGLLVPSSTSDGE 123
SV + R+C ++ G L+ S SDGE
Sbjct 518 SVVYCRNLDRLCCCSRQGGLLFYSLSDGE 546
> YKR031c
Length=1683
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 27/49 (55%), Gaps = 2/49 (4%)
Query 38 SSLVNKMAIRTHFESSNEVGVFGKL-TNSYCLVASGGSEHFYSAIEAEL 85
SS++ KM+ T + N G F + TNS+C G ++F+S EA L
Sbjct 655 SSII-KMSTSTPWSKPNRFGSFAPVRTNSFCKFLVDGRDYFWSLSEALL 702
> At5g63520
Length=529
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 17/61 (27%), Positives = 30/61 (49%), Gaps = 2/61 (3%)
Query 69 VASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIGRVCVGNKHGLLVPSSTSDGELQQLR 128
+ G E + I +G +P++ + V G ++G+ +K G + STSD EL +
Sbjct 111 ITCGNMEETLTLITERVGSRVPIIVSVVTG--ILGKEACNDKAGEVRLHSTSDDELFDVA 168
Query 129 N 129
N
Sbjct 169 N 169
> At5g13950
Length=978
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 117 SSTSDGELQQLRNSLPDGVSVRRVEERL 144
S SDGE LR LP+GV V +V + L
Sbjct 149 SCLSDGERNYLRQFLPEGVDVEQVVQAL 176
> At2g44230
Length=542
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 60 GKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIGR 104
GKL Y SGGS S IE + G + PV +AS+ G A+ +
Sbjct 385 GKLHRMYLSQHSGGSWADASEIEFQGGGNKPVAYASLNGHAMYSK 429
> At1g12620
Length=621
Score = 27.7 bits (60), Expect = 7.3, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 23/52 (44%), Gaps = 2/52 (3%)
Query 5 SCCCCCCFLRSATVTFGPIFLCGIIFRPGWAAFSSLVNKMAIRTHFESSNEV 56
+CCC C L A G I G + P FS+L+N + + + E+
Sbjct 115 NCCCRCRKLSLAFSAMGKIIKLG--YEPDTVTFSTLINGLCLEGRVSEALEL 164
Lambda K H
0.325 0.138 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40