bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1873_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
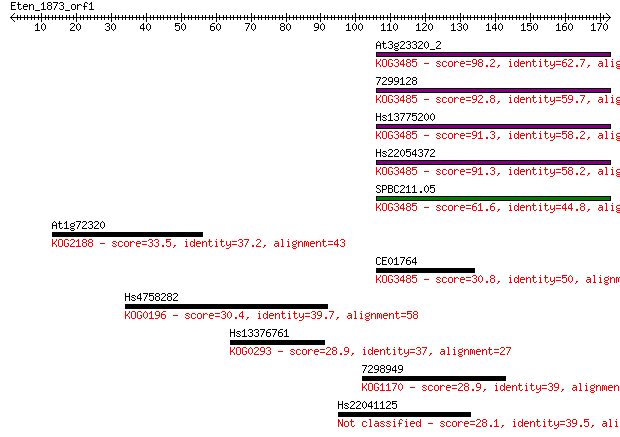
At3g23320_2 98.2 7e-21
7299128 92.8 3e-19
Hs13775200 91.3 8e-19
Hs22054372 91.3 9e-19
SPBC211.05 61.6 8e-10
At1g72320 33.5 0.20
CE01764 30.8 1.4
Hs4758282 30.4 1.7
Hs13376761 28.9 5.0
7298949 28.9 5.6
Hs22041125 28.1 8.8
> At3g23320_2
Length=84
Score = 98.2 bits (243), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 42/67 (62%), Positives = 55/67 (82%), Gaps = 0/67 (0%)
Query 106 DRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAENESIR 165
DRF I+SQL+HLQ+KY GTG+AD +R +W +NIQRD+ AS++GHY L+YFA+AENESI
Sbjct 2 DRFNINSQLEHLQAKYVGTGHADLSRFEWTVNIQRDSYASYIGHYPMLSYFAIAENESIG 61
Query 166 RIRYRCL 172
R RY +
Sbjct 62 RERYNFM 68
> 7299128
Length=85
Score = 92.8 bits (229), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 40/67 (59%), Positives = 53/67 (79%), Gaps = 0/67 (0%)
Query 106 DRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAENESIR 165
+R+ IHSQL+HLQSKY GTG+ADTT+ +W N RD+LAS++GHY L YFA+AENES
Sbjct 3 ERYNIHSQLEHLQSKYIGTGHADTTKFEWLTNQHRDSLASYMGHYDILNYFAIAENESKA 62
Query 166 RIRYRCL 172
R+R+ +
Sbjct 63 RVRFNLM 69
> Hs13775200
Length=86
Score = 91.3 bits (225), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 39/67 (58%), Positives = 53/67 (79%), Gaps = 0/67 (0%)
Query 106 DRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAENESIR 165
DR+TIHSQL+HLQSKY GTG+ADTT+ +W +N RD+ S++GH+ L YFA+AENES
Sbjct 3 DRYTIHSQLEHLQSKYIGTGHADTTKWEWLVNQHRDSYCSYMGHFDLLNYFAIAENESKA 62
Query 166 RIRYRCL 172
R+R+ +
Sbjct 63 RVRFNLM 69
> Hs22054372
Length=121
Score = 91.3 bits (225), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 39/67 (58%), Positives = 53/67 (79%), Gaps = 0/67 (0%)
Query 106 DRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAENESIR 165
DR+TIHSQL+HLQSKY GTG+ADTT+ +W +N RD+ S++GH+ L YFA+AENES
Sbjct 3 DRYTIHSQLEHLQSKYIGTGHADTTKWEWLVNQHRDSYCSYMGHFDLLNYFAIAENESKA 62
Query 166 RIRYRCL 172
R+R+ +
Sbjct 63 RVRFNLM 69
> SPBC211.05
Length=85
Score = 61.6 bits (148), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 30/67 (44%), Positives = 41/67 (61%), Gaps = 0/67 (0%)
Query 106 DRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAENESIR 165
DR ++L+ LQ++Y G GNA TT+ +W +N RDTL+S VGH LAY A A E
Sbjct 3 DRLRSQAKLEQLQARYVGVGNAFTTKYEWMVNQHRDTLSSVVGHPPLLAYMATALGEPRV 62
Query 166 RIRYRCL 172
++R L
Sbjct 63 QVRKNLL 69
> At1g72320
Length=753
Score = 33.5 bits (75), Expect = 0.20, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 27/45 (60%), Gaps = 2/45 (4%)
Query 13 QGPFSARKTTAAAAATTPEQQRRQQQHQQ--YNRCCSDFSSSENS 55
QGP+ RK A+ P+Q + +Q+ +Q YN CS F S++++
Sbjct 613 QGPYLLRKLDIDGYASRPDQWKSRQEAKQSTYNEFCSAFGSNKSN 657
> CE01764
Length=87
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 21/29 (72%), Gaps = 1/29 (3%)
Query 106 DRFTIHSQLQHLQSKYPGTG-NADTTRLD 133
+RF + +QL+HLQSKY GT + +R+D
Sbjct 6 ERFHVLAQLEHLQSKYTGTAMRHEPSRMD 34
> Hs4758282
Length=998
Score = 30.4 bits (67), Expect = 1.7, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 32/71 (45%), Gaps = 13/71 (18%)
Query 34 RRQQQHQQYNRCCS-DFSSSENSTR-----------SCPPCLCLTWRPAIPVNLLFACRK 81
+ Q Q +RC + FS E S+R S PP + T P+ P NL+F +
Sbjct 285 KSSSQDLQCSRCPTHSFSDKEGSSRCECEDGYYRAPSDPPYVACTRPPSAPQNLIFNINQ 344
Query 82 GLLLL-WSPAA 91
+ L WSP A
Sbjct 345 TTVSLEWSPPA 355
> Hs13376761
Length=367
Score = 28.9 bits (63), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 15/27 (55%), Gaps = 0/27 (0%)
Query 64 CLTWRPAIPVNLLFACRKGLLLLWSPA 90
C++W P IP + A G + +W PA
Sbjct 324 CVSWNPQIPSMMASASDDGTVRIWGPA 350
> 7298949
Length=1301
Score = 28.9 bits (63), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 21/41 (51%), Gaps = 1/41 (2%)
Query 102 MSNMDRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDT 142
+S +DRF +H Q Q + GTGN L WG + DT
Sbjct 309 LSEIDRFNMHKQCQ-VAVMPLGTGNDLARVLGWGSSCDDDT 348
> Hs22041125
Length=439
Score = 28.1 bits (61), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 95 LLLQQAKMSNMDRFTIHSQLQHLQSKYPGTGNADTTRL 132
LL + K+S + I S LQ +K P T + DTT+L
Sbjct 400 LLRHRLKLSTSEVVRIQSALQAFNAKLPNTMDYDTTKL 437
Lambda K H
0.322 0.131 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2562785186
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40