bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1817_orf2
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
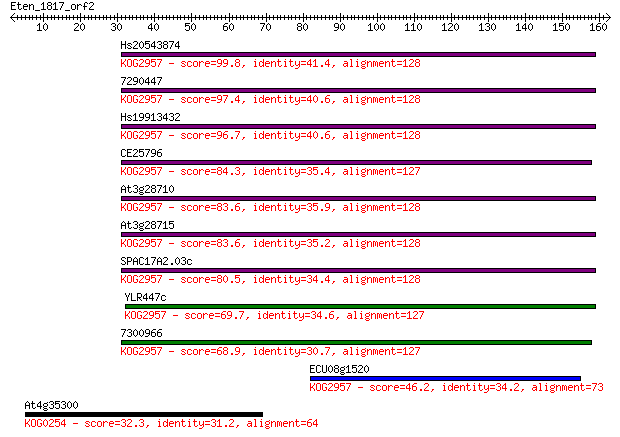
Hs20543874 99.8 2e-21
7290447 97.4 1e-20
Hs19913432 96.7 2e-20
CE25796 84.3 1e-16
At3g28710 83.6 2e-16
At3g28715 83.6 2e-16
SPAC17A2.03c 80.5 1e-15
YLR447c 69.7 2e-12
7300966 68.9 4e-12
ECU08g1520 46.2 3e-05
At4g35300 32.3 0.43
> Hs20543874
Length=350
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 53/128 (41%), Positives = 73/128 (57%), Gaps = 13/128 (10%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR L+P FG LYP+G + +A + ++ + Y Y +++
Sbjct 236 DRETLYPTFGKLYPEGLRLLAQAEDFDQMKNVADHYGVYKPLFEAVGG------------ 283
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
S KTLED+ Y VQM L F +QFHYGVFYA+VKLKEQEIRNIVWIAE I + +
Sbjct 284 -SGGKTLEDVFYEREVQMNVLAFNRQFHYGVFYAYVKLKEQEIRNIVWIAECISQRHRTK 342
Query 151 IDAIVPLF 158
I++ +P+
Sbjct 343 INSYIPIL 350
> 7290447
Length=350
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 52/128 (40%), Positives = 72/128 (56%), Gaps = 12/128 (9%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR L+PN G +YPDG + +A + ++ E Y Y ++D G + D
Sbjct 235 DRAKLYPNCGKMYPDGLAALARADDYEQVKTVAEYYAEYAALFD-------GSGNNPGD- 286
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
KTLED + V++ F QQFH+GVFYA++KLKEQE RNIVWIAE + K +
Sbjct 287 ----KTLEDKFFEHEVKLDVYAFLQQFHFGVFYAYLKLKEQECRNIVWIAECVAQKHRAK 342
Query 151 IDAIVPLF 158
ID +P+F
Sbjct 343 IDNYIPIF 350
> Hs19913432
Length=351
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 52/128 (40%), Positives = 74/128 (57%), Gaps = 12/128 (9%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR LFP+ G LYP+G ++ +A + ++ + YP Y + + G + D
Sbjct 236 DRAKLFPHCGRLYPEGLAQLARADDYEQVKNVADYYPEY-------KLLFEGAGSNPGD- 287
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
KTLED + V++ +L F QFH+GVFYA+VKLKEQE RNIVWIAE I + +
Sbjct 288 ----KTLEDRFFEHEVKLNKLAFLNQFHFGVFYAFVKLKEQECRNIVWIAECIAQRHRAK 343
Query 151 IDAIVPLF 158
ID +P+F
Sbjct 344 IDNYIPIF 351
> CE25796
Length=343
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 45/127 (35%), Positives = 69/127 (54%), Gaps = 12/127 (9%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR L+P G L+PDG + +A + ++ E Y Y + + G +
Sbjct 228 DRQKLYPRCGKLFPDGLTGLSRADDYDQVKQVCEFYSDY-------KPLFEGSGNGPGE- 279
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
KTLED + V++ ++ QFH+GVFYA++KLKEQE+RNI+WIAE I + +
Sbjct 280 ----KTLEDKFFEHEVKLNVHSYLHQFHFGVFYAFIKLKEQEMRNIIWIAECISQRHRTK 335
Query 151 IDAIVPL 157
ID +P+
Sbjct 336 IDNYIPI 342
> At3g28710
Length=351
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 46/129 (35%), Positives = 72/129 (55%), Gaps = 14/129 (10%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR L+ NFG LYP G + + + +R +E YP Y ++ + G+
Sbjct 236 DRKKLYSNFGLLYPYGHEELAICEDIDQVRGVMEKYPPYQAIFSK---MSYGES------ 286
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
+ L+ Y E V+ L FEQQFHY VF+A+++L+EQEIRN++WI+E + +K
Sbjct 287 ----QMLDKAFYEEEVRRLCLAFEQQFHYAVFFAYMRLREQEIRNLMWISECVAQNQKSR 342
Query 151 I-DAIVPLF 158
I D++V +F
Sbjct 343 IHDSVVYMF 351
> At3g28715
Length=351
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 45/129 (34%), Positives = 70/129 (54%), Gaps = 14/129 (10%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR L+ NFG LYP G + + + +R +E YP Y ++ +
Sbjct 236 DRKKLYSNFGLLYPYGHEELAICEDIDQVRGVMEKYPPYQAIFSKMSY------------ 283
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
+ L+ Y E V+ L FEQQFHY VF+A+++L+EQEIRN++WI+E + +K
Sbjct 284 -GESQMLDKAFYEEEVRRLCLAFEQQFHYAVFFAYMRLREQEIRNLMWISECVAQNQKSR 342
Query 151 I-DAIVPLF 158
I D++V +F
Sbjct 343 IHDSVVYMF 351
> SPAC17A2.03c
Length=343
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 44/128 (34%), Positives = 69/128 (53%), Gaps = 16/128 (12%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
+R ++P+FG LYP T + +A N + A Y + +DQ
Sbjct 232 ERAKMYPSFGRLYPFSTSILARAENAGDVENACSLVKEYSDFFDQ--------------- 276
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
+ K+L+D + V++ +L F QQFHYG+ YA++KL+EQEIRN+ WIAE I +++
Sbjct 277 -NSQKSLDDHFNEKEVELNKLAFLQQFHYGIVYAFLKLREQEIRNLTWIAECISQNQRDR 335
Query 151 IDAIVPLF 158
IVP+
Sbjct 336 ALNIVPIL 343
> YLR447c
Length=345
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 44/127 (34%), Positives = 63/127 (49%), Gaps = 19/127 (14%)
Query 32 RHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADLT 91
+ L PN G LYP T + +A + +RAAL NVY+ R F
Sbjct 238 KSDLLPNIGKLYPLATFHLAQAQDFEGVRAALA------NVYEY-RGFL----------- 279
Query 92 SRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEHI 151
LED Y+ +++C F QQF +AW+K KEQE+RNI WIAE I ++E I
Sbjct 280 -ETGNLEDHFYQLEMELCRDAFTQQFAISTVWAWMKSKEQEVRNITWIAECIAQNQRERI 338
Query 152 DAIVPLF 158
+ + ++
Sbjct 339 NNYISVY 345
> 7300966
Length=350
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 39/127 (30%), Positives = 64/127 (50%), Gaps = 11/127 (8%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
+R +FP GYL + + +R + Y ++D ERD +
Sbjct 232 ERLKMFPTCGYLPKIALASMSTLNDTDKIRDVCNVFDGYGKMFDNL-------ERDSDGM 284
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
TLED + TF QQ+H+G+FY+++KLK+ E+RNIVWI+E I ++ +
Sbjct 285 I----TLEDRFLMMEAKKNVQTFLQQYHFGIFYSFIKLKQLEVRNIVWISECIAQRQTDR 340
Query 151 IDAIVPL 157
I+A +P+
Sbjct 341 INAFIPI 347
> ECU08g1520
Length=341
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 46/76 (60%), Gaps = 3/76 (3%)
Query 82 GDERDKADLTSRF---KTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVW 138
G E K+ L++R+ K + ++L R +Q + +F Y++ K+KEQEI+NI+W
Sbjct 263 GLEDMKSVLSTRYNFDKDISNVLIRAELQKYQESFSMYGDLSCVYSYFKIKEQEIKNILW 322
Query 139 IAEMILMKRKEHIDAI 154
+AE I+ R++ +D +
Sbjct 323 VAECIIQNRRDAMDHL 338
> At4g35300
Length=729
Score = 32.3 bits (72), Expect = 0.43, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 31/65 (47%), Gaps = 1/65 (1%)
Query 5 RRLSSVKALLQVLLQQQQQQQQQQQLDRHALFPNFGYLYPDGTDRIR-KAWNDPTLRAAL 63
RR S+ L L ++ R ALFP+FG ++ G ++ R + W++ L
Sbjct 291 RRQGSLIDPLVTLFGSVHEKMPDTGSMRSALFPHFGSMFSVGGNQPRHEDWDEENLVGEG 350
Query 64 EPYPS 68
E YPS
Sbjct 351 EDYPS 355
Lambda K H
0.325 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2244926132
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40