bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1814_orf1
Length=362
Score E
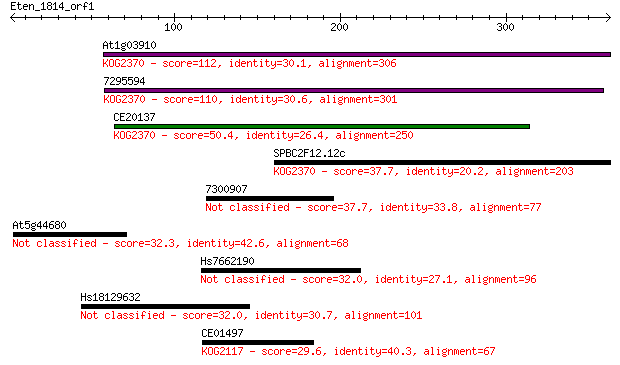
Sequences producing significant alignments: (Bits) Value
At1g03910 112 2e-24
7295594 110 6e-24
CE20137 50.4 5e-06
SPBC2F12.12c 37.7 0.035
7300907 37.7 0.038
At5g44680 32.3 1.5
Hs7662190 32.0 1.9
Hs18129632 32.0 2.0
CE01497 29.6 9.3
> At1g03910
Length=672
Score = 112 bits (279), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 92/307 (29%), Positives = 153/307 (49%), Gaps = 28/307 (9%)
Query 57 LRAERKKLLSEHFGYSNDNNPFGDRLLAEPFVWKKK-NLLLQAAGKKHATTVNALLTTSV 115
LRA +K GYSND+NPFGD L E FVW+KK + +V A
Sbjct 103 LRAAKKLKTQSVSGYSNDSNPFGDSNLTETFVWRKKIEKDVHRGVPLEEFSVKAEKRRHR 162
Query 116 SKVQEIKSVKKRREEREAEEQLMQAQREELQREKEREHFQDYFEKEEVFLRQMHMKKTEI 175
++ E++ VKKRREER E+ + + L RE+ R F D+ +KEE F ++EI
Sbjct 163 ERMTEVEKVKKRREERAVEKARHEEEMALLARERARAEFHDWEKKEEEFHFDQSKVRSEI 222
Query 176 RAEQNREQPIDFIVKGLRMLNGERFEKVSVLPVPPHEVFDCFNKEPMTVQMIEEMLSDVK 235
R + R +PID + K L+G + + + P+ VF + +TV+ +EE+ D+K
Sbjct 223 RLREGRLKPIDVLCK---HLDGS--DDLDIELSEPYMVF-----KGLTVKDMEELRDDIK 272
Query 236 LHIGVDTTAVDKDYQDFWQALLVLTKDALERAERKQKVLEELRNNSSADAAAQAAAILAN 295
+++ +D + +W+AL+V+ L A RK+ L+ R A +LA
Sbjct 273 MYLDLDRATPTR--VQYWEALIVVCDWELAEA-RKRDALDRAR----VRGEEPPAELLAQ 325
Query 296 NTSVGDVRDADSGIAVGVTSDITAILSGKSPEELDALKEGIKLKLETEEDVDTAYWESIL 355
+ G+ GV +D+ +L GK+ EL L+ I+ +L + YWE++L
Sbjct 326 ----------ERGLHAGVEADVRKLLDGKTHAELVELQLDIESQLRSGSAKVVEYWEAVL 375
Query 356 QKVPLFR 362
+++ +++
Sbjct 376 KRLEIYK 382
> 7295594
Length=720
Score = 110 bits (274), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 92/303 (30%), Positives = 151/303 (49%), Gaps = 37/303 (12%)
Query 58 RAERKKLLSEHFGYSNDNNPFGDRLLAEPFVWKKKNLLLQAAGKKHATTVNAL-LTTSVS 116
R ER +E+ YSN++NPFGD L F W KK L ++ TV L L +
Sbjct 154 RRERMGWDNEYQTYSNEDNPFGDSNLTSTFHWGKK-LEVEGLSNLSTKTVEVLSLQKQLE 212
Query 117 KVQEIKSVKKRREEREAEEQLMQAQREELQREKEREHFQDYFEKEEVFLRQMHMKKTEIR 176
+E++ VKKRR+ERE E Q+ + QR KE F+++ +E+ F + ++EIR
Sbjct 213 NRRELEKVKKRRQERELERQVREDDLMMQQRAKEAVQFREWQRQEDQFHLEQARLRSEIR 272
Query 177 AEQNREQPIDFIVKGLRMLNGERFEKVSVLPVPPHEVFDCFNKEPMTVQMIEEMLSDVKL 236
R +PID + + + N E E+ L + HE + N P V+ +E++L D+K+
Sbjct 273 IRDGRAKPIDLLAQYVAAGN-EPLEE--CLEMQMHEPYVLLNGLP--VEELEDLLVDIKV 327
Query 237 HIGVDTTAVDKDYQDFWQALLVLTKDALERAERKQKVLEELRNNSSADAAAQAAAILANN 296
+ ++ + DFW ++ + +D L+R Q+ LE A N
Sbjct 328 YEELEQGK----HIDFWNDMITIVQDELQR----QQKLE------------------AEN 361
Query 297 TSVGDVRDADSGIAVGVTSDITAILSGKSPEELDALKEGIKLKLETEED-VDTAYWESIL 355
+S+ RD GI V D+ I GK+ ++L+ ++ I+ K+ D VD +YWES+L
Sbjct 362 SSLNQRRD---GIHQAVVKDVADIFRGKNAQQLEEMRHRIEAKISGRADGVDISYWESLL 418
Query 356 QKV 358
++
Sbjct 419 SQL 421
> CE20137
Length=667
Score = 50.4 bits (119), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 66/270 (24%), Positives = 112/270 (41%), Gaps = 34/270 (12%)
Query 64 LLSEHFGYSNDNNPFGDRLLAEPFVWKKKNLLLQAAGKKHATTVNALLTTSV---SKVQE 120
L+ Y+N NNPF D L + FVW KK L+ GK T TS + E
Sbjct 83 LIPPELNYTNLNNPFNDTKLTQTFVWGKK---LEREGKSGLTQDEITKQTSQRIRKNLHE 139
Query 121 IKSVKKRREEREAEEQLMQAQREELQREKEREHFQDYFEKEEVFLRQMHMKKTEIRAEQN 180
K+ R+ R A ++ M E ++R+ + Q KE F ++T IR +Q
Sbjct 140 AAEFKRIRDSRAAAKEDM----EMMKRDADLRAGQISDTKEREFQMDQIKERTRIRIDQG 195
Query 181 REQPIDFIVKGLRMLNGERFEKVSVLPVPPHEV---FDCFNKEPMTVQMIEEMLSDVKLH 237
R + ID + + R + E +P E+ + +V E+++ D+K +
Sbjct 196 RAKAIDLLSRYARFAD----ENPHTAKIPDFELENPMEYLKASCKSVDDYEDLIEDIKTY 251
Query 238 IGVDTTAVDKDYQDFWQALLVLTKDALE------RAERKQKVLEELRN---NSSADAAAQ 288
VD A + + +W + + +D ++ R + V E++N N S D +
Sbjct 252 REVDGWAKN---ETWWMDVTRIAEDEIQKKAAQNRGDVHASVQTEVQNMFKNKSIDELLK 308
Query 289 A-----AAILANNTSVGDVRDADSGIAVGV 313
A I N+ + G +D D + V +
Sbjct 309 LEDQMDAKIRGNSGNKGYWQDLDDQLKVFI 338
> SPBC2F12.12c
Length=517
Score = 37.7 bits (86), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 41/204 (20%), Positives = 84/204 (41%), Gaps = 35/204 (17%)
Query 160 KEEVFLRQMHMKKTEIRAEQNREQPIDFIVKGLRMLNGERFEKVSVLPVPPHEVFDCFNK 219
+E FLR +++ IR + +R +PID ++ + ++ G +E+ D F+
Sbjct 62 QEREFLRLQQLRRAIIRLKNDRSKPIDKLLVSVYLMPGPEWEENGEKNSIDFGTVDMFDP 121
Query 220 EPMTVQMIEEMLSDVKLHIG-VDTTAVDKDYQDFWQALLVLTKDALERAERKQKVLEELR 278
+ E L ++ +IG + ++ +W+ + +L +E
Sbjct 122 LSVIQSYKAEDLEEISRNIGDIQQLETNQCRLTYWKYVKMLVNSRME------------- 168
Query 279 NNSSADAAAQAAAILANNTSVGDVRDADSGIAVGVTSDITAILSGKSPEELDALKEGIKL 338
+ VG G+ V V ++ IL+ KS E+L+ L+ I+
Sbjct 169 -----------------HNKVGQF---SRGLKV-VAGEVQRILAPKSFEQLEQLEAQIQK 207
Query 339 KLETEEDVDTAYWESILQKVPLFR 362
KL++ +DT YW +L + ++
Sbjct 208 KLKSNVPLDTDYWVDLLNSLKSYK 231
> 7300907
Length=775
Score = 37.7 bits (86), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 45/83 (54%), Gaps = 14/83 (16%)
Query 119 QEIKSVKKRREEREAEEQLMQAQREELQREKEREHFQDYFEKEEVFLRQMHMKKTEIR-- 176
Q+ KS++ + E ++Q Q+ R +H + + +KE VF+ Q+ K TEI
Sbjct 270 QDYKSMQMQMAEESPQDQDRQSLR--------CKHTESFLQKERVFIAQLIAKATEISPT 321
Query 177 -AEQNREQPID---FIVKGLRML 195
+ ++ P+D +IVKGLR+L
Sbjct 322 IFDNGKKGPVDMNAYIVKGLRLL 344
> At5g44680
Length=353
Score = 32.3 bits (72), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 37/73 (50%), Gaps = 9/73 (12%)
Query 3 KPTKMAKRRRRSSSSEREDRRAPPNSSEG-----PNAKGTSQPGSVAAARPVKPVTVQEL 57
KP K RSSS++ + +P NS G P QPGS+AAAR + QE
Sbjct 76 KPAGSCKELLRSSSTKSKPVISPENSDGGYKEVMPMVIVQKQPGSIAAARREEVAMKQE- 134
Query 58 RAERKKLLSEHFG 70
ERKK +S H+G
Sbjct 135 --ERKKKIS-HYG 144
> Hs7662190
Length=1533
Score = 32.0 bits (71), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 49/104 (47%), Gaps = 9/104 (8%)
Query 116 SKVQEIKSVKKRREEREAEEQLMQAQREELQREKEREHFQDYFEKEEVFLRQMHMKKTEI 175
+KV + + + +E + Q + Q+ +QR++E+ H D+ ++ VF+ Q H++ E
Sbjct 187 AKVNSVTELLSKLQETDKHLQRVTEQQTSIQRKQEKLHCHDHEKQMNVFMEQ-HIRHLEK 245
Query 176 RAEQNREQPIDFIVKGLR--------MLNGERFEKVSVLPVPPH 211
+Q + FI L+ M + EK SV P P+
Sbjct 246 LQQQQIDIQTHFISAALKTSSFQPVSMPSSRAVEKYSVKPEHPN 289
> Hs18129632
Length=202
Score = 32.0 bits (71), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 31/116 (26%), Positives = 55/116 (47%), Gaps = 18/116 (15%)
Query 44 AAARPVKPVTVQELRAERKKLLSEHFGYSNDNNPFGDRLLAEPFV---------WKKKN- 93
A++RP+K + + +L S+H+G + + G L P V W KK+
Sbjct 74 ASSRPIKLLQQPGTDTPQGRLYSDHYGLYHTSPSLGG--LTRPVVLWSQQDVCKWLKKHC 131
Query 94 ----LLLQAAGKKHATTVNALLTTSVSKVQEIKSVKKRREEREAEEQLMQAQ-REE 144
L+ A +HA T ALL + K+Q + + + + +E +Q+++ Q REE
Sbjct 132 PHNYLVYVEAFSQHAITGRALLRLNAEKLQRM-GLAQEAQRQEVLQQVLRLQVREE 186
> CE01497
Length=392
Score = 29.6 bits (65), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 40/75 (53%), Gaps = 9/75 (12%)
Query 117 KVQEIKSVKKRREEREAEE--------QLMQAQREELQREKEREHFQDYFEKEEVFLRQM 168
K +E + K RE + AE QL Q REE Q+ +ERE D F+ +EVF+
Sbjct 87 KKEEARKADKNRESKYAENIIKAHARRQLEQFSREERQQLREREKEGDEFDDKEVFVTGA 146
Query 169 HMKKTEIRAEQNREQ 183
+ K+ E +++REQ
Sbjct 147 YRKQQE-EVKKHREQ 160
Lambda K H
0.312 0.128 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8839980072
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40