bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1810_orf1
Length=205
Score E
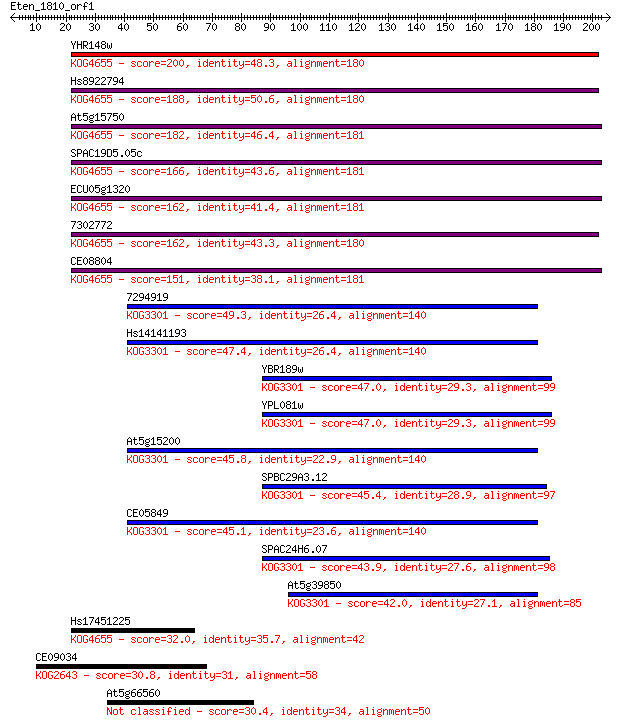
Sequences producing significant alignments: (Bits) Value
YHR148w 200 1e-51
Hs8922794 188 7e-48
At5g15750 182 3e-46
SPAC19D5.05c 166 2e-41
ECU05g1320 162 3e-40
7302772 162 4e-40
CE08804 151 1e-36
7294919 49.3 6e-06
Hs14141193 47.4 2e-05
YBR189w 47.0 3e-05
YPL081w 47.0 3e-05
At5g15200 45.8 6e-05
SPBC29A3.12 45.4 8e-05
CE05849 45.1 1e-04
SPAC24H6.07 43.9 2e-04
At5g39850 42.0 9e-04
Hs17451225 32.0 0.81
CE09034 30.8 1.8
At5g66560 30.4 2.8
> YHR148w
Length=183
Score = 200 bits (509), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 87/181 (48%), Positives = 131/181 (72%), Gaps = 1/181 (0%)
Query 22 MRQLKYHEQRLLRKVNFFEWKRDKTARENKFLKKYLIQDREDYHRYNKLCGLITKLVAGL 81
+R+LK+HEQ+LL+KV+F EWK+D+ R+ + ++ Y IQ+REDYH+YN++CG I +L L
Sbjct 2 VRKLKHHEQKLLKKVDFLEWKQDQGHRDTQVMRTYHIQNREDYHKYNRICGDIRRLANKL 61
Query 82 RKLPPEDSFRMKMTELLLDKLYRMGVVSRREGLGAVEG-LAASAFCRRRLPVVLLRLRMA 140
LPP D FR K +LLLDKLY MGV++ + + +E + SA CRRRLPV++ RL+MA
Sbjct 62 SLLPPTDPFRRKHEQLLLDKLYAMGVLTTKSKISDLENKVTVSAICRRRLPVIMHRLKMA 121
Query 141 THLQQAVEYVEQGHVRLGAEVVTSPSLHITRDAEDHLAWAEGSAIKRHVAAFRSQADDFD 200
+Q AV+++EQGHVR+G ++ P+ +TR+ ED++ W + S IK+ + +R+Q DDFD
Sbjct 122 ETIQDAVKFIEQGHVRVGPNLINDPAYLVTRNMEDYVTWVDNSKIKKTLLRYRNQIDDFD 181
Query 201 L 201
Sbjct 182 F 182
> Hs8922794
Length=184
Score = 188 bits (477), Expect = 7e-48, Method: Compositional matrix adjust.
Identities = 91/181 (50%), Positives = 124/181 (68%), Gaps = 1/181 (0%)
Query 22 MRQLKYHEQRLLRKVNFFEWK-RDKTARENKFLKKYLIQDREDYHRYNKLCGLITKLVAG 80
+R+LK+HEQ+LL++V+F W+ D E + L++Y +Q REDY RYN+L + +L
Sbjct 2 VRKLKFHEQKLLKQVDFLNWEVTDHNLHELRVLRRYRLQRREDYTRYNQLSRAVRELARR 61
Query 81 LRKLPPEDSFRMKMTELLLDKLYRMGVVSRREGLGAVEGLAASAFCRRRLPVVLLRLRMA 140
LR LP D FR++ + LLDKLY +G+V R L + + AS+FCRRRLP VLL+LRMA
Sbjct 62 LRDLPERDQFRVRASAALLDKLYALGLVPTRGSLELCDFVTASSFCRRRLPTVLLKLRMA 121
Query 141 THLQQAVEYVEQGHVRLGAEVVTSPSLHITRDAEDHLAWAEGSAIKRHVAAFRSQADDFD 200
HLQ AV +VEQGHVR+G +VVT P+ +TR ED + W + S IKRHV + + DDFD
Sbjct 122 QHLQAAVAFVEQGHVRVGPDVVTDPAFLVTRSMEDFVTWVDSSKIKRHVLEYNEERDDFD 181
Query 201 L 201
L
Sbjct 182 L 182
> At5g15750
Length=194
Score = 182 bits (463), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 84/193 (43%), Positives = 124/193 (64%), Gaps = 12/193 (6%)
Query 22 MRQLKYHEQRLLRKVNFFEWKRDKTARENKFLKKYLIQDREDYHRYNKLCGL-------- 73
MR+LKYHE++L++KVNF EWKR+ REN+ +Y + R+DY + L L
Sbjct 1 MRKLKYHEKKLIKKVNFLEWKREGNHRENEITYRYHMGSRDDYKKLVPLFALKALFYLFF 60
Query 74 ----ITKLVAGLRKLPPEDSFRMKMTELLLDKLYRMGVVSRREGLGAVEGLAASAFCRRR 129
+ KL ++++ P D FR++MT++LL+KLY MGV+ R+ L E L+ S+FCRRR
Sbjct 61 FFWMVQKLTNIMKQMDPADPFRIQMTDMLLEKLYNMGVIPTRKSLTLTERLSVSSFCRRR 120
Query 130 LPVVLLRLRMATHLQQAVEYVEQGHVRLGAEVVTSPSLHITRDAEDHLAWAEGSAIKRHV 189
L VL+ L+ A H ++AV Y+EQGHVR+G E +T P+ +TR+ ED + W + S IKR V
Sbjct 121 LSTVLVHLKFAEHHKEAVTYIEQGHVRVGPETITDPAFLVTRNMEDFITWVDSSKIKRKV 180
Query 190 AAFRSQADDFDLL 202
+ DD+D+L
Sbjct 181 LEYNDTLDDYDML 193
> SPAC19D5.05c
Length=183
Score = 166 bits (421), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 79/183 (43%), Positives = 117/183 (63%), Gaps = 2/183 (1%)
Query 22 MRQLKYHEQRLLRKVNFFEWKRDKTA-RENKFLKKYLIQDREDYHRYNKLCGLITKLVAG 80
MR LK+HEQ+LLRKV+F +K D R+ +++Y I RE+Y +YN +CG +L
Sbjct 1 MRILKHHEQKLLRKVDFLNYKNDDNNHRDVMVMRRYHISKREEYQKYNIICGKFRQLAHR 60
Query 81 LRKLPPEDSFRMKMTELLLDKLYRMGVVSRREGLGAVEGLA-ASAFCRRRLPVVLLRLRM 139
L L P D FR++ LLL+KL+ MG++ + + +E A SA CRRRLPV++ +LRM
Sbjct 61 LSLLDPTDPFRLQYENLLLEKLFDMGILPSKSKMSDIENKANVSAICRRRLPVIMCKLRM 120
Query 140 ATHLQQAVEYVEQGHVRLGAEVVTSPSLHITRDAEDHLAWAEGSAIKRHVAAFRSQADDF 199
+ + +A +EQGHVR+G V+T P+ +TR ED + W + S IKR +A + + DD+
Sbjct 121 SQVVSEATRLIEQGHVRVGPHVITDPAYLVTRSLEDFVTWTDTSKIKRTIAKYNDKLDDY 180
Query 200 DLL 202
DLL
Sbjct 181 DLL 183
> ECU05g1320
Length=180
Score = 162 bits (411), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 75/181 (41%), Positives = 119/181 (65%), Gaps = 1/181 (0%)
Query 22 MRQLKYHEQRLLRKVNFFEWKRDKTARENKFLKKYLIQDREDYHRYNKLCGLITKLVAGL 81
MR+LK+HE++LL++VNF EWK TARE+ + KY ++DRE+Y +Y+++ G + KL L
Sbjct 1 MRELKFHEKKLLKQVNFLEWKNTNTAREHLAIAKYGLRDREEYTKYSRIVGKVRKLAEAL 60
Query 82 RKLPPEDSFRMKMTELLLDKLYRMGVVSRREGLGAVEGLAASAFCRRRLPVVLLRLRMAT 141
+L D R +T+ L+ +LY +GV+ R+ + + + S+FC RRLP+V+ R+RM
Sbjct 61 ARLKESDPVRTTITKKLVGRLYSVGVIKNRKLVDCTK-VTVSSFCERRLPMVMKRIRMVP 119
Query 142 HLQQAVEYVEQGHVRLGAEVVTSPSLHITRDAEDHLAWAEGSAIKRHVAAFRSQADDFDL 201
+ A +VE GHVRLG++VV PS I++ ED ++W + S IKR + F + DDF+
Sbjct 120 SFKDATRFVEHGHVRLGSKVVYDPSTIISKAMEDFVSWVDKSKIKRKIDEFNGELDDFNY 179
Query 202 L 202
+
Sbjct 180 V 180
> 7302772
Length=200
Score = 162 bits (410), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 78/199 (39%), Positives = 121/199 (60%), Gaps = 19/199 (9%)
Query 22 MRQLKYHEQRLLRKVNFFEWKRDKTARENKFLKKYLIQDREDYHRYNKLCGLITKLVAGL 81
+R+LK+HEQ+LL+KV+F WK D +ENK L+++ IQ REDY +YNKL I +L +
Sbjct 2 VRKLKFHEQKLLKKVDFITWKVDNGGKENKILRRFHIQKREDYTKYNKLSREIRELAERI 61
Query 82 RKLPPEDSFRMKMTELLLDKLYRMGVVSRREGLGAVEGLAASAFCRRRLPVVLLR----- 136
KL + F+ + T +LL+KL+ MGV + + L ++AS FCRRRLPV++++
Sbjct 62 AKLDASEPFKTEATTMLLNKLHAMGVSNDQLTLETAAKISASHFCRRRLPVIMVKREYHP 121
Query 137 --------------LRMATHLQQAVEYVEQGHVRLGAEVVTSPSLHITRDAEDHLAWAEG 182
+RM+ HL+ A + +E GHVR+G E++ P+ ++R+ ED + W +G
Sbjct 122 LISTSFSYNHFPLAVRMSEHLKAATDLIEHGHVRVGPEMIKDPAFLVSRNLEDFVTWVDG 181
Query 183 SAIKRHVAAFRSQADDFDL 201
S IK HV + DDF +
Sbjct 182 SKIKEHVLRYNDMRDDFQM 200
> CE08804
Length=183
Score = 151 bits (381), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 69/181 (38%), Positives = 110/181 (60%), Gaps = 0/181 (0%)
Query 22 MRQLKYHEQRLLRKVNFFEWKRDKTARENKFLKKYLIQDREDYHRYNKLCGLITKLVAGL 81
+R+LK HEQ+LL+K +F W+ D+ ++ L+K+ ++ RE Y YN L ++ +
Sbjct 2 VRKLKTHEQKLLKKTDFMSWQVDQQGKQGDMLRKFYVKKREHYALYNTLAAKSREVADLI 61
Query 82 RKLPPEDSFRMKMTELLLDKLYRMGVVSRREGLGAVEGLAASAFCRRRLPVVLLRLRMAT 141
+ L D FR K TE +L K Y G+V + L + + ++F RRRLPVV+ + M
Sbjct 62 KNLSESDPFRSKCTEDMLTKFYAAGLVPTSDTLERIGKVTGASFARRRLPVVMRNIGMCE 121
Query 142 HLQQAVEYVEQGHVRLGAEVVTSPSLHITRDAEDHLAWAEGSAIKRHVAAFRSQADDFDL 201
++ A + VEQGHVR+G ++VT P+ +TR +ED + W + S IK+HV + + DDFDL
Sbjct 122 SVKTASDLVEQGHVRIGTKLVTDPAFMVTRSSEDMITWTKASKIKKHVMDYNNTRDDFDL 181
Query 202 L 202
+
Sbjct 182 M 182
> 7294919
Length=195
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 37/141 (26%), Positives = 68/141 (48%), Gaps = 1/141 (0%)
Query 41 WKRDKTARENKFLKKYLIQDREDYHRYNKLCGLITKLVAGLRKLPPEDSFRMKMTELLLD 100
+++ + +E K + +Y ++++ + R I K L L +D R+ LL
Sbjct 21 YEKARLDQELKIIGEYGLRNKREVWRVKYALAKIRKAARELLTLDEKDEKRLFQGNALLR 80
Query 101 KLYRMGVVSR-REGLGAVEGLAASAFCRRRLPVVLLRLRMATHLQQAVEYVEQGHVRLGA 159
+L R+GV+ R L V GL F RRL + +L +A + A + Q H+R+
Sbjct 81 RLVRIGVLDESRMKLDYVLGLKIEDFLERRLQTQVFKLGLAKSIHHARVLIRQRHIRVRK 140
Query 160 EVVTSPSLHITRDAEDHLAWA 180
+VV PS + D++ H+ ++
Sbjct 141 QVVNIPSFVVRLDSQKHIDFS 161
> Hs14141193
Length=194
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 37/143 (25%), Positives = 69/143 (48%), Gaps = 5/143 (3%)
Query 41 WKRDKTARENKFLKKYLIQDREDYHRYNKLCGLITKLVAGLRKLPPEDSFRMKMTELLLD 100
+++ + +E K + +Y ++++ + R I K L L +D R+ LL
Sbjct 20 FEKSRLDQELKLIGEYGLRNKREVWRVKFTLAKIRKAARELLTLDEKDPRRLFEGNALLR 79
Query 101 KLYRMGVVSRREG---LGAVEGLAASAFCRRRLPVVLLRLRMATHLQQAVEYVEQGHVRL 157
+L R+GV+ EG L + GL F RRL + +L +A + A + Q H+R+
Sbjct 80 RLVRIGVLD--EGKMKLDYILGLKIEDFLERRLQTQVFKLGLAKSIHHARVLIRQRHIRV 137
Query 158 GAEVVTSPSLHITRDAEDHLAWA 180
+VV PS + D++ H+ ++
Sbjct 138 RKQVVNIPSFIVRLDSQKHIDFS 160
> YBR189w
Length=195
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 50/100 (50%), Gaps = 1/100 (1%)
Query 87 EDSFRMKMTELLLDKLYRMGVVSR-REGLGAVEGLAASAFCRRRLPVVLLRLRMATHLQQ 145
+D R+ L+ +L R+GV+S ++ L V L F RRL + +L +A +
Sbjct 65 KDPKRLFEGNALIRRLVRVGVLSEDKKKLDYVLALKVEDFLERRLQTQVYKLGLAKSVHH 124
Query 146 AVEYVEQGHVRLGAEVVTSPSLHITRDAEDHLAWAEGSAI 185
A + Q H+ +G ++V PS + D+E H+ +A S
Sbjct 125 ARVLITQRHIAVGKQIVNIPSFMVRLDSEKHIDFAPTSPF 164
> YPL081w
Length=197
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 50/100 (50%), Gaps = 1/100 (1%)
Query 87 EDSFRMKMTELLLDKLYRMGVVSR-REGLGAVEGLAASAFCRRRLPVVLLRLRMATHLQQ 145
+D R+ L+ +L R+GV+S ++ L V L F RRL + +L +A +
Sbjct 65 KDPKRLFEGNALIRRLVRVGVLSEDKKKLDYVLALKVEDFLERRLQTQVYKLGLAKSVHH 124
Query 146 AVEYVEQGHVRLGAEVVTSPSLHITRDAEDHLAWAEGSAI 185
A + Q H+ +G ++V PS + D+E H+ +A S
Sbjct 125 ARVLITQRHIAVGKQIVNIPSFMVRLDSEKHIDFAPTSPF 164
> At5g15200
Length=198
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 32/141 (22%), Positives = 66/141 (46%), Gaps = 1/141 (0%)
Query 41 WKRDKTARENKFLKKYLIQDREDYHRYNKLCGLITKLVAGLRKLPPEDSFRMKMTELLLD 100
+++++ E K + +Y ++++ + R I L L + R+ E LL
Sbjct 21 YEKERLDSELKLVGEYGLRNKRELWRVQYSLSRIRNAARDLLTLDEKSPRRIFEGEALLR 80
Query 101 KLYRMGVVSRREG-LGAVEGLAASAFCRRRLPVVLLRLRMATHLQQAVEYVEQGHVRLGA 159
++ R G++ + L V L F RRL ++ + MA + + + Q H+R+G
Sbjct 81 RMNRYGLLDESQNKLDYVLALTVENFLERRLQTIVFKSGMAKSIHHSRVLIRQRHIRVGK 140
Query 160 EVVTSPSLHITRDAEDHLAWA 180
++V PS + D++ H+ +A
Sbjct 141 QLVNIPSFMVRLDSQKHIDFA 161
> SPBC29A3.12
Length=192
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 49/98 (50%), Gaps = 1/98 (1%)
Query 87 EDSFRMKMTELLLDKLYRMGVVSR-REGLGAVEGLAASAFCRRRLPVVLLRLRMATHLQQ 145
+D R+ ++ +L R+G++ R L V L F RRL + +L +A +
Sbjct 65 KDPKRLFEGNAIIRRLVRLGILDESRMKLDYVLALRIEDFLERRLQTQVFKLGLAKSIHH 124
Query 146 AVEYVEQGHVRLGAEVVTSPSLHITRDAEDHLAWAEGS 183
A + Q H+R+G ++V PS + DA+ H+ +A S
Sbjct 125 ARVLIFQRHIRVGKQIVNVPSFVVRLDAQKHIDFALSS 162
> CE05849
Length=189
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/141 (23%), Positives = 69/141 (48%), Gaps = 1/141 (0%)
Query 41 WKRDKTARENKFLKKYLIQDREDYHRYNKLCGLITKLVAGLRKLPPEDSFRMKMTELLLD 100
+++++ +E K + + ++++ + R + K L L +D R+ LL
Sbjct 19 FEKERLDQELKLIGTFGLKNKREVWRVKYTLAKVRKAARELLTLEDKDPKRLFEGNALLR 78
Query 101 KLYRMGVVSR-REGLGAVEGLAASAFCRRRLPVVLLRLRMATHLQQAVEYVEQGHVRLGA 159
+L ++GV+ + L V GL F RRL + +L +A + A ++Q H+R+
Sbjct 79 RLVKIGVLDETKMKLDYVLGLKVEDFLERRLQTQVFKLGLAKSIHHARILIKQHHIRVRR 138
Query 160 EVVTSPSLHITRDAEDHLAWA 180
+VV PS + D++ H+ ++
Sbjct 139 QVVDVPSFIVRLDSQKHIDFS 159
> SPAC24H6.07
Length=191
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 48/99 (48%), Gaps = 1/99 (1%)
Query 87 EDSFRMKMTELLLDKLYRMGVVSR-REGLGAVEGLAASAFCRRRLPVVLLRLRMATHLQQ 145
+D R+ ++ +L R+G++ R L V L F RRL + +L +A +
Sbjct 65 KDPKRLFEGNAIIRRLVRLGILDETRMKLDYVLALRIEDFLERRLQTQVFKLGLAKSIHH 124
Query 146 AVEYVEQGHVRLGAEVVTSPSLHITRDAEDHLAWAEGSA 184
A + Q H+R+G ++V PS + D + H+ +A S
Sbjct 125 ARVLIFQRHIRVGKQIVNVPSFVVRLDTQKHIDFALSSP 163
> At5g39850
Length=197
Score = 42.0 bits (97), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 44/86 (51%), Gaps = 1/86 (1%)
Query 96 ELLLDKLYRMGVVSRREG-LGAVEGLAASAFCRRRLPVVLLRLRMATHLQQAVEYVEQGH 154
E LL ++ R G++ + L V L F RRL ++ + MA + A + Q H
Sbjct 76 EALLRRMNRYGLLDETQNKLDYVLALTVENFLERRLQTIVFKSGMAKSIHHARVLIRQRH 135
Query 155 VRLGAEVVTSPSLHITRDAEDHLAWA 180
+R+G ++V PS + +++ H+ ++
Sbjct 136 IRVGRQLVNIPSFMVRVESQKHVDFS 161
> Hs17451225
Length=109
Score = 32.0 bits (71), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 24/43 (55%), Gaps = 1/43 (2%)
Query 22 MRQLKYHEQRLLRKVNFFEWK-RDKTARENKFLKKYLIQDRED 63
+R+LK+H Q LL++V+F W+ D E Y ++ ED
Sbjct 2 VRKLKFHVQNLLKQVDFLNWEVTDHNLHELCIQWHYQLKQWED 44
> CE09034
Length=431
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 31/62 (50%), Gaps = 4/62 (6%)
Query 10 GTAAAAAAAAAEMRQLKYHEQRLLRKVNFFEWKRDKTAREN----KFLKKYLIQDREDYH 65
G A + E +Q + Q L ++ F+E+ R KTA + + +Y I + +DYH
Sbjct 245 GLRGNATLSFDEFQQFYENLQEELMEIEFYEFARGKTAISPVDFARLILRYSIVNFDDYH 304
Query 66 RY 67
+Y
Sbjct 305 KY 306
> At5g66560
Length=668
Score = 30.4 bits (67), Expect = 2.8, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Query 34 RKVNFFEWKRDKTARENKFLKKYLIQDREDYHRYNKLCGLITKLVAGLRK 83
R+V+ +WK KT REN+ L+ + R HR + C + K++A + K
Sbjct 576 RQVDAGKWK--KTVRENQVLRLDMDTMRTRVHRLERECSNMKKVIAKIDK 623
Lambda K H
0.322 0.133 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3658712052
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40