bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1787_orf1
Length=110
Score E
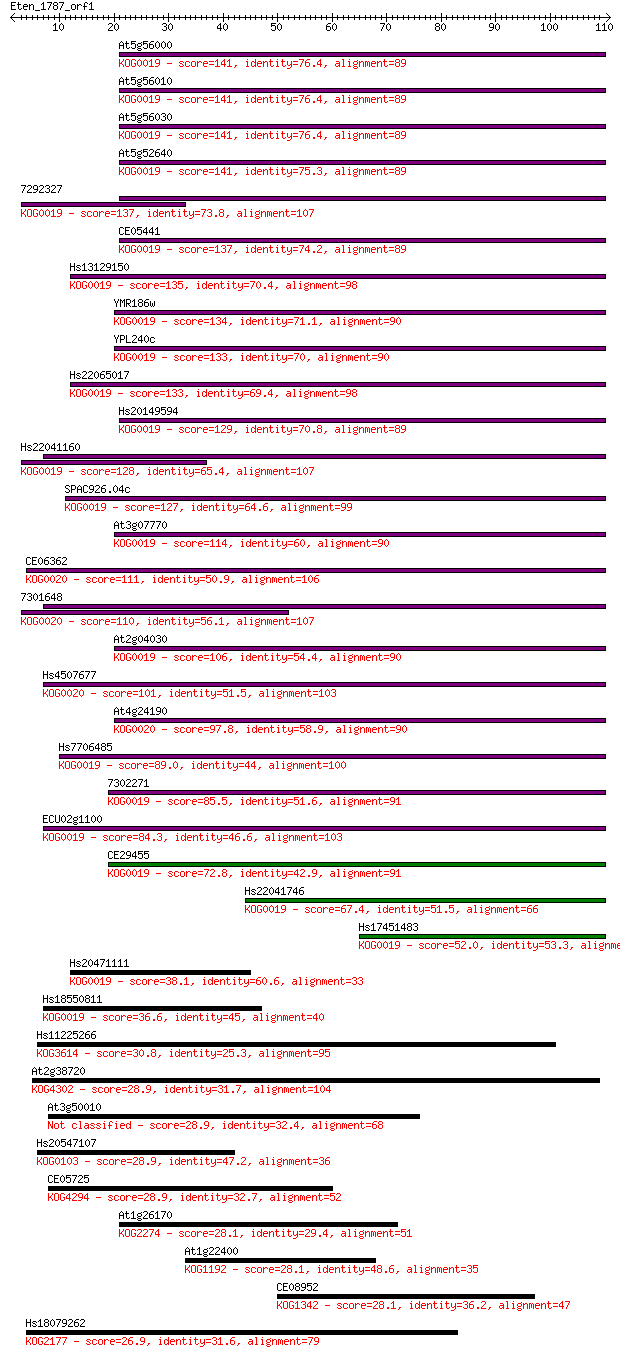
Sequences producing significant alignments: (Bits) Value
At5g56000 141 2e-34
At5g56010 141 2e-34
At5g56030 141 3e-34
At5g52640 141 3e-34
7292327 137 4e-33
CE05441 137 6e-33
Hs13129150 135 1e-32
YMR186w 134 3e-32
YPL240c 133 8e-32
Hs22065017 133 1e-31
Hs20149594 129 1e-30
Hs22041160 128 3e-30
SPAC926.04c 127 5e-30
At3g07770 114 3e-26
CE06362 111 3e-25
7301648 110 5e-25
At2g04030 106 1e-23
Hs4507677 101 4e-22
At4g24190 97.8 4e-21
Hs7706485 89.0 2e-18
7302271 85.5 2e-17
ECU02g1100 84.3 5e-17
CE29455 72.8 2e-13
Hs22041746 67.4 6e-12
Hs17451483 52.0 3e-07
Hs20471111 38.1 0.004
Hs18550811 36.6 0.011
Hs11225266 30.8 0.62
At2g38720 28.9 2.3
At3g50010 28.9 2.3
Hs20547107 28.9 2.5
CE05725 28.9 2.6
At1g26170 28.1 4.0
At1g22400 28.1 4.2
CE08952 28.1 4.3
Hs18079262 26.9 9.9
> At5g56000
Length=699
Score = 141 bits (356), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 68/89 (76%), Positives = 80/89 (89%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++T+ KL +PELFI +I
Sbjct 9 FQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLDGQPELFIHII 68
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PDK NNTLTI DSGIGMTKA+LVNNLGT+
Sbjct 69 PDKTNNTLTIIDSGIGMTKADLVNNLGTI 97
> At5g56010
Length=699
Score = 141 bits (356), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 68/89 (76%), Positives = 80/89 (89%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++T+ KL +PELFI +I
Sbjct 9 FQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLDGQPELFIHII 68
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PDK NNTLTI DSGIGMTKA+LVNNLGT+
Sbjct 69 PDKTNNTLTIIDSGIGMTKADLVNNLGTI 97
> At5g56030
Length=699
Score = 141 bits (356), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 68/89 (76%), Positives = 80/89 (89%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++T+ KL +PELFI +I
Sbjct 9 FQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLDGQPELFIHII 68
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PDK NNTLTI DSGIGMTKA+LVNNLGT+
Sbjct 69 PDKTNNTLTIIDSGIGMTKADLVNNLGTI 97
> At5g52640
Length=705
Score = 141 bits (356), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 67/89 (75%), Positives = 81/89 (91%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QL+SLIINTFYSNKEIFLRELISN+SDALDKIR+E++T+ KL +PELFIRL+
Sbjct 14 FQAEINQLLSLIINTFYSNKEIFLRELISNSSDALDKIRFESLTDKSKLDGQPELFIRLV 73
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PDK+N TL+I DSGIGMTKA+LVNNLGT+
Sbjct 74 PDKSNKTLSIIDSGIGMTKADLVNNLGTI 102
> 7292327
Length=717
Score = 137 bits (346), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 66/89 (74%), Positives = 79/89 (88%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QLMSLIINTFYSNKEIFLRELISNASDALDKIRYE++T+P KL + EL+I+LI
Sbjct 10 FQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLDSGKELYIKLI 69
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P+K TLTI D+GIGMTK++LVNNLGT+
Sbjct 70 PNKTAGTLTIIDTGIGMTKSDLVNNLGTI 98
Score = 30.4 bits (67), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 17/30 (56%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLI 32
KEGLE+ E E EKKK EE KA + L L+
Sbjct 531 KEGLELPEDESEKKKREEDKAKFESLCKLM 560
> CE05441
Length=702
Score = 137 bits (344), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 66/89 (74%), Positives = 78/89 (87%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QLMSLIINTFYSNKEI+LRELISNASDALDKIRY+A+TEP +L T ELFI++
Sbjct 10 FQAEIAQLMSLIINTFYSNKEIYLRELISNASDALDKIRYQALTEPSELDTGKELFIKIT 69
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P+K TLTI D+GIGMTKA+LVNNLGT+
Sbjct 70 PNKEEKTLTIMDTGIGMTKADLVNNLGTI 98
> Hs13129150
Length=732
Score = 135 bits (341), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 69/98 (70%), Positives = 83/98 (84%), Gaps = 1/98 (1%)
Query 12 EEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKT 71
EEE + F +A+I QLMSLIINTFYSNKEIFLRELISN+SDALDKIRYE++T+P KL +
Sbjct 14 EEEVETFA-FQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLDS 72
Query 72 KPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
EL I LIP+K + TLTI D+GIGMTKA+L+NNLGT+
Sbjct 73 GKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTI 110
> YMR186w
Length=705
Score = 134 bits (338), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 64/90 (71%), Positives = 78/90 (86%), Gaps = 0/90 (0%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRL 79
E +A+I QLMSLIINT YSNKEIFLRELISNASDALDKIRY+A+++P++L+T+P+LFIR+
Sbjct 7 EFQAEITQLMSLIINTVYSNKEIFLRELISNASDALDKIRYQALSDPKQLETEPDLFIRI 66
Query 80 IPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P L I DSGIGMTKAEL+NNLGT+
Sbjct 67 TPKPEEKVLEIRDSGIGMTKAELINNLGTI 96
> YPL240c
Length=709
Score = 133 bits (335), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 63/90 (70%), Positives = 78/90 (86%), Gaps = 0/90 (0%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRL 79
E +A+I QLMSLIINT YSNKEIFLRELISNASDALDKIRY+++++P++L+T+P+LFIR+
Sbjct 7 EFQAEITQLMSLIINTVYSNKEIFLRELISNASDALDKIRYKSLSDPKQLETEPDLFIRI 66
Query 80 IPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P L I DSGIGMTKAEL+NNLGT+
Sbjct 67 TPKPEQKVLEIRDSGIGMTKAELINNLGTI 96
> Hs22065017
Length=343
Score = 133 bits (334), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 68/98 (69%), Positives = 82/98 (83%), Gaps = 1/98 (1%)
Query 12 EEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKT 71
EEE + F +A+I QLMSLIINTFYSNKEIFLRELISN+SDALDKI YE++T+P KL +
Sbjct 14 EEEVETFA-FQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIWYESLTDPSKLDS 72
Query 72 KPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
EL I LIP+K + TLTI D+GIGMTKA+L+NNLGT+
Sbjct 73 GKELHINLIPNKQDQTLTIVDTGIGMTKADLINNLGTI 110
> Hs20149594
Length=724
Score = 129 bits (325), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 63/89 (70%), Positives = 76/89 (85%), Gaps = 0/89 (0%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRLI 80
+A+I QLMSLIINTFYSNKEIFLRELISNASDALDKIRYE++T+P KL + EL I +I
Sbjct 17 FQAEIAQLMSLIINTFYSNKEIFLRELISNASDALDKIRYESLTDPSKLDSGKELKIDII 76
Query 81 PDKANNTLTIEDSGIGMTKAELVNNLGTL 109
P+ TLT+ D+GIGMTKA+L+NNLGT+
Sbjct 77 PNPQERTLTLVDTGIGMTKADLINNLGTI 105
> Hs22041160
Length=343
Score = 128 bits (321), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 66/103 (64%), Positives = 83/103 (80%), Gaps = 1/103 (0%)
Query 7 EIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEP 66
E+ E+E + F +A+I QLMSLIINTFYSNKEIFL ELISNASDALDKIRYE++T+P
Sbjct 4 EVHLGEKEVETFA-FQAEIAQLMSLIINTFYSNKEIFLWELISNASDALDKIRYESLTDP 62
Query 67 EKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
KL + EL I +IP+ +TLT+ D+GIGMTKA+L+NNLGT+
Sbjct 63 SKLDSGKELKIDIIPNTQEHTLTLVDTGIGMTKADLINNLGTI 105
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLIINTF 36
KEGLE+ E EEKK+ EE KA + L + T
Sbjct 273 KEGLELPEDGEEKKRMEERKAKFENLCKFMKETL 306
> SPAC926.04c
Length=704
Score = 127 bits (319), Expect = 5e-30, Method: Compositional matrix adjust.
Identities = 64/99 (64%), Positives = 78/99 (78%), Gaps = 3/99 (3%)
Query 11 SEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLK 70
S E KFE A+I QLMSLIINT YSNKEIFLRELISNASDALDKIRY+++++P L
Sbjct 2 SNTETFKFE---AEISQLMSLIINTVYSNKEIFLRELISNASDALDKIRYQSLSDPHALD 58
Query 71 TKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
+ +LFIR+ PDK N L+I D+GIGMTK +L+NNLG +
Sbjct 59 AEKDLFIRITPDKENKILSIRDTGIGMTKNDLINNLGVI 97
> At3g07770
Length=803
Score = 114 bits (286), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 54/90 (60%), Positives = 71/90 (78%), Gaps = 0/90 (0%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRL 79
E +A++ +LM LI+N+ YSNKE+FLRELISNASDALDK+RY ++T PE K P+L IR+
Sbjct 98 EYQAEVSRLMDLIVNSLYSNKEVFLRELISNASDALDKLRYLSVTNPELSKDAPDLDIRI 157
Query 80 IPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
DK N +T+ DSGIGMT+ ELV+ LGT+
Sbjct 158 YADKENGIITLTDSGIGMTRQELVDCLGTI 187
> CE06362
Length=760
Score = 111 bits (278), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 54/108 (50%), Positives = 81/108 (75%), Gaps = 2/108 (1%)
Query 4 EGLEIDESEEEKKKFE--ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYE 61
+GL + + +E + K E E +A++ ++M LIIN+ Y NKEIFLRELISNASDALDKIR
Sbjct 47 DGLSVSQIKELRSKAEKHEFQAEVNRMMKLIINSLYRNKEIFLRELISNASDALDKIRLL 106
Query 62 AITEPEKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
++T+PE+L+ E+ +++ D+ N L I D+G+GMT+ +L+NNLGT+
Sbjct 107 SLTDPEQLRETEEMSVKIKADRENRLLHITDTGVGMTRQDLINNLGTI 154
> 7301648
Length=787
Score = 110 bits (276), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 57/103 (55%), Positives = 76/103 (73%), Gaps = 1/103 (0%)
Query 7 EIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEP 66
++ E E+ KKF + ++ ++M LIIN+ Y NKEIFLRELISNASDA+DKIR A++
Sbjct 64 QLKEIREKAKKFT-FQTEVNRMMKLIINSLYRNKEIFLRELISNASDAIDKIRLLALSNS 122
Query 67 EKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
++L+T PEL IR+ DK N L I DSGIGMT +L+NNLGT+
Sbjct 123 KELETNPELHIRIKADKENKALHIMDSGIGMTHQDLINNLGTI 165
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 29/49 (59%), Gaps = 7/49 (14%)
Query 3 KEGLEIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNA 51
KEG +++ESE+ KK FE LK+ + L+ + ++ L++ IS A
Sbjct 590 KEGFQLNESEKSKKNFESLKSTFEPLVKWL-------NDVALKDQISKA 631
> At2g04030
Length=780
Score = 106 bits (264), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 49/90 (54%), Positives = 71/90 (78%), Gaps = 0/90 (0%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIRL 79
E +A++ +L+ LI+++ YS+KE+FLREL+SNASDALDK+R+ ++TEP L +L IR+
Sbjct 80 EYQAEVSRLLDLIVHSLYSHKEVFLRELVSNASDALDKLRFLSVTEPSLLGDGGDLEIRI 139
Query 80 IPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PD N T+TI D+GIGMTK EL++ LGT+
Sbjct 140 KPDPDNGTITITDTGIGMTKEELIDCLGTI 169
> Hs4507677
Length=803
Score = 101 bits (251), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 53/103 (51%), Positives = 73/103 (70%), Gaps = 1/103 (0%)
Query 7 EIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEP 66
+I E E+ +KF +A++ ++M LIIN+ Y NKEIFLRELISNASDALDKIR ++T+
Sbjct 65 QIRELREKSEKFA-FQAEVNRMMKLIINSLYKNKEIFLRELISNASDALDKIRLISLTDE 123
Query 67 EKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
L EL +++ DK N L + D+G+GMT+ ELV NLGT+
Sbjct 124 NALSGNEELTVKIKCDKEKNLLHVTDTGVGMTREELVKNLGTI 166
> At4g24190
Length=823
Score = 97.8 bits (242), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 53/94 (56%), Positives = 70/94 (74%), Gaps = 6/94 (6%)
Query 20 ELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKL----KTKPEL 75
E +A++ +LM +IIN+ YSNK+IFLRELISNASDALDKIR+ A+T+ + L K E+
Sbjct 80 EFQAEVSRLMDIIINSLYSNKDIFLRELISNASDALDKIRFLALTDKDVLGEGDTAKLEI 139
Query 76 FIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
I+L DKA L+I D GIGMTK +L+ NLGT+
Sbjct 140 QIKL--DKAKKILSIRDRGIGMTKEDLIKNLGTI 171
> Hs7706485
Length=704
Score = 89.0 bits (219), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 44/100 (44%), Positives = 71/100 (71%), Gaps = 3/100 (3%)
Query 10 ESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKL 69
ES + E +A+ ++L+ ++ + YS KE+F+RELISNASDAL+K+R++ +++ + L
Sbjct 79 ESVQGSTSKHEFQAETKKLLDIVARSLYSEKEVFIRELISNASDALEKLRHKLVSDGQAL 138
Query 70 KTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
PE+ I L + T+TI+D+GIGMT+ ELV+NLGT+
Sbjct 139 ---PEMEIHLQTNAEKGTITIQDTGIGMTQEELVSNLGTI 175
> 7302271
Length=691
Score = 85.5 bits (210), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 47/94 (50%), Positives = 67/94 (71%), Gaps = 4/94 (4%)
Query 19 EELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITE-PEKL--KTKPEL 75
E +A+ +QL+ ++ + YS+ E+F+RELISNASDAL+K RY +++ E L K +P L
Sbjct 66 HEFQAETRQLLDIVARSLYSDHEVFVRELISNASDALEKFRYTSLSAGGENLAGKDRP-L 124
Query 76 FIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
IR+ DK L I+D+GIGMTK ELV+NLGT+
Sbjct 125 EIRITTDKPLMQLIIQDTGIGMTKEELVSNLGTI 158
> ECU02g1100
Length=690
Score = 84.3 bits (207), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 48/106 (45%), Positives = 68/106 (64%), Gaps = 6/106 (5%)
Query 7 EIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEP 66
+I + E FE D+ Q+M +I + YS+KE+FLREL+SN+SDA DK++
Sbjct 12 KIKDKHSETHGFE---VDVNQMMDTMIKSVYSSKELFLRELVSNSSDACDKLKALYFQLR 68
Query 67 EK---LKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
EK L L I +IP+K N TLTI+D+GIGMTK +L+N +GT+
Sbjct 69 EKGCVLDPVTSLGIEIIPNKDNRTLTIKDNGIGMTKPDLMNFIGTI 114
> CE29455
Length=657
Score = 72.8 bits (177), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 39/91 (42%), Positives = 58/91 (63%), Gaps = 3/91 (3%)
Query 19 EELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKTKPELFIR 78
E +A+ + LM ++ + YS+ E+F+RELISNASDAL+K RY + + P IR
Sbjct 30 HEFQAETRNLMDIVAKSLYSHSEVFVRELISNASDALEKRRYAELK--GDVAEGPSE-IR 86
Query 79 LIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
+ +K T+T ED+GIGM + +LV LGT+
Sbjct 87 ITTNKDKRTITFEDTGIGMNREDLVKFLGTI 117
> Hs22041746
Length=1595
Score = 67.4 bits (163), Expect = 6e-12, Method: Composition-based stats.
Identities = 34/66 (51%), Positives = 47/66 (71%), Gaps = 0/66 (0%)
Query 44 LRELISNASDALDKIRYEAITEPEKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELV 103
L E + NA+ L + E++T+P KL + E I LIP+K + TLTI D+GIGMTKA+L+
Sbjct 635 LAETMPNANLILPCLLPESLTDPSKLDSGKEPHISLIPNKQDRTLTIVDTGIGMTKADLI 694
Query 104 NNLGTL 109
NNLGT+
Sbjct 695 NNLGTI 700
> Hs17451483
Length=112
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/45 (53%), Positives = 32/45 (71%), Gaps = 0/45 (0%)
Query 65 EPEKLKTKPELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGTL 109
+ KL + L I L+P+K N TLTI D+ IGMTK +L+NNLGT+
Sbjct 2 DSSKLDSGKGLHINLVPNKQNQTLTIVDTRIGMTKTDLINNLGTV 46
> Hs20471111
Length=217
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/33 (60%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 12 EEEKKKFEELKADIQQLMSLIINTFYSNKEIFL 44
EEE+ + +A I + MSLIINTFY NKEIFL
Sbjct 4 EEEEVEICTFQALIAEPMSLIINTFYFNKEIFL 36
> Hs18550811
Length=329
Score = 36.6 bits (83), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 29/40 (72%), Gaps = 1/40 (2%)
Query 7 EIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRE 46
E+ + +EE + F +A++ Q M L INTFYSN++IFL++
Sbjct 4 EVYQGKEEVETFA-FQAEVAQFMYLNINTFYSNQKIFLQD 42
> Hs11225266
Length=1165
Score = 30.8 bits (68), Expect = 0.62, Method: Composition-based stats.
Identities = 24/96 (25%), Positives = 46/96 (47%), Gaps = 6/96 (6%)
Query 6 LEIDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITE 65
++I +SE E D++++M ++ SNK F+R + N +D D + Y + E
Sbjct 361 VDIAKSEIFNGDVEWKSCDLEEVM---VDALVSNKPEFVRLFVDNGADVADFLTYGRLQE 417
Query 66 PEKLKTKPELFIRLIPDKANNT-LTIEDSGIGMTKA 100
+ ++ L L+ K LT+ +G+G +A
Sbjct 418 LYRSVSRKSLLFDLLQRKQEEARLTL--AGLGTQQA 451
> At2g38720
Length=587
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 33/120 (27%), Positives = 57/120 (47%), Gaps = 19/120 (15%)
Query 5 GLEIDESEEEKKKFEELKADIQQLMS------LIINTFYSNKEIFLRELISNASDALDKI 58
G E+DES+ ++K +EL+AD+Q L + +N++ S L + S AL+ +
Sbjct 140 GSEVDESDLTQRKLDELRADLQDLRNEKAVRLQKVNSYISAVHELSEILSFDFSKALNSV 199
Query 59 RYEAITEPEKLKTKP----------ELFIRLIPDKANNTLTIEDSGIGMTKAELVNNLGT 108
+ ++TE K +K EL L +K L ++ G+G + EL N + T
Sbjct 200 -HSSLTEFSKTHSKSISNDTLARFTELVKSLKAEKHERLLKLQ--GLGRSMQELWNLMET 256
> At3g50010
Length=769
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 37/68 (54%), Gaps = 5/68 (7%)
Query 8 IDESEEEKKKFEELKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPE 67
+D + EEK + +EL IQ+++SL+ + ++ L ELIS S + I E
Sbjct 115 MDLTTEEKPESDELVLLIQEILSLVNSRDSDSQPKRLVELISFISQ-----KISLIDETL 169
Query 68 KLKTKPEL 75
++K +PEL
Sbjct 170 EVKPEPEL 177
> Hs20547107
Length=840
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 17/36 (47%), Positives = 21/36 (58%), Gaps = 2/36 (5%)
Query 6 LEIDESEEEKKKFEELKADIQQLMSLIINTFYSNKE 41
+ ESEE K FEEL IQQ M +I + + NKE
Sbjct 687 IRFQESEERPKLFEELGKQIQQYMKII--SSFKNKE 720
> CE05725
Length=840
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 8 IDESEEEKKKFEEL-KADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIR 59
+DES +F+EL K D ++SL N+F S ++F+ + D+IR
Sbjct 141 LDESSSNSSRFDELDKFDDVDILSLYRNSFQSADDLFVSCEFGKSGSCQDEIR 193
> At1g26170
Length=931
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 31/51 (60%), Gaps = 1/51 (1%)
Query 21 LKADIQQLMSLIINTFYSNKEIFLRELISNASDALDKIRYEAITEPEKLKT 71
+++++ + S + +TF S+ +++LR I A D+ D RY++ E + L T
Sbjct 258 IESELMGMFSPLWHTFESSLQVYLRSSIDGAEDSYDG-RYDSDGEEKSLDT 307
> At1g22400
Length=489
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 21/37 (56%), Gaps = 3/37 (8%)
Query 33 INTFYSNKEIFLRELISNASDALDKIRYEAITE--PE 67
+NT Y N FLR SNA D L R+E+I + PE
Sbjct 45 VNTVY-NHNRFLRSRGSNALDGLPSFRFESIADGLPE 80
> CE08952
Length=461
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query 50 NASDALDKIRYEAITEPEKLKTKPELFIRLIPDKANNTLTIEDSGIG 96
N+SD L K++ + I E+L P + +R IP+ A + L +DS I
Sbjct 358 NSSDMLAKLQTDVIANLEQLTFVPSVQMRPIPEDALSALN-DDSLIA 403
> Hs18079262
Length=488
Score = 26.9 bits (58), Expect = 9.9, Method: Composition-based stats.
Identities = 25/86 (29%), Positives = 41/86 (47%), Gaps = 8/86 (9%)
Query 4 EGLEIDESEEEKKK----FEELKAD-IQQLMSL--IINTFYSNKEIFLRELISNASDALD 56
E E+ + E+E+KK EE + D + Q SL +I+ + EL+ + SD +
Sbjct 198 EQRELKKLEQEEKKGLRIIEEAENDLVHQTQSLRELISDLERRCQGSTMELLQDVSDVTE 257
Query 57 KIRYEAITEPEKLKTKPELFIRLIPD 82
+ + + +PE L TK R PD
Sbjct 258 RSEFWTLRKPEALPTKLRSMFR-APD 282
Lambda K H
0.312 0.133 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1195973986
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40