bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1784_orf1
Length=152
Score E
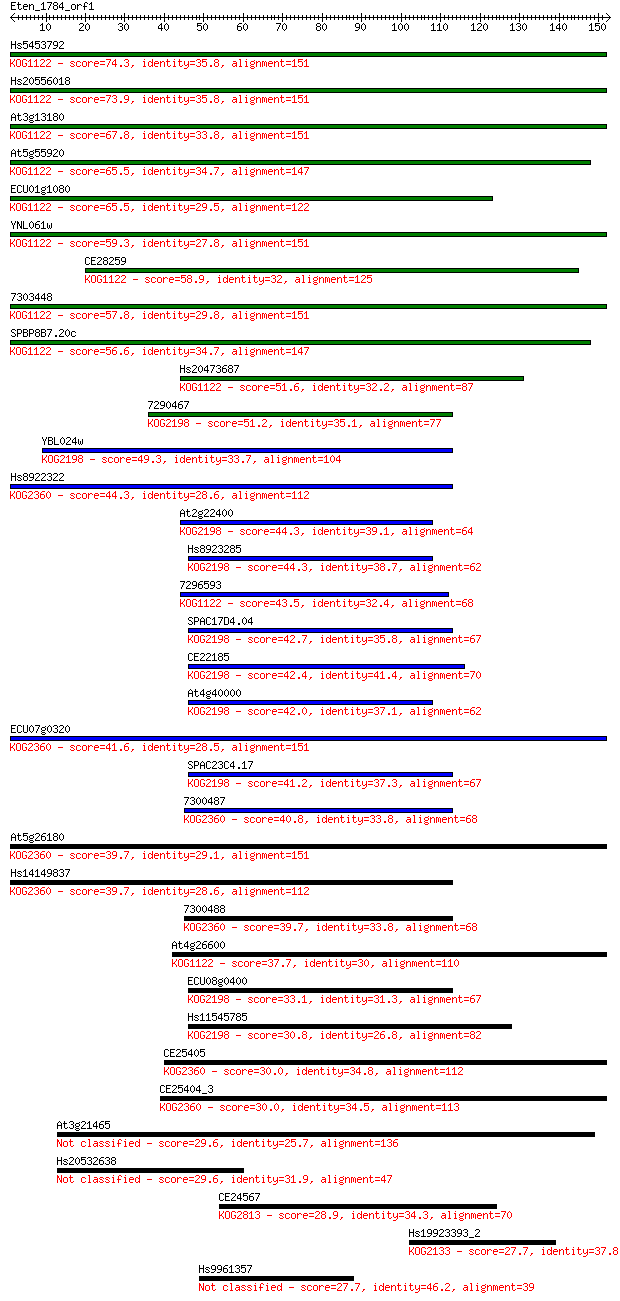
Sequences producing significant alignments: (Bits) Value
Hs5453792 74.3 8e-14
Hs20556018 73.9 1e-13
At3g13180 67.8 7e-12
At5g55920 65.5 4e-11
ECU01g1080 65.5 4e-11
YNL061w 59.3 3e-09
CE28259 58.9 4e-09
7303448 57.8 7e-09
SPBP8B7.20c 56.6 2e-08
Hs20473687 51.6 6e-07
7290467 51.2 8e-07
YBL024w 49.3 3e-06
Hs8922322 44.3 9e-05
At2g22400 44.3 9e-05
Hs8923285 44.3 1e-04
7296593 43.5 2e-04
SPAC17D4.04 42.7 2e-04
CE22185 42.4 4e-04
At4g40000 42.0 4e-04
ECU07g0320 41.6 6e-04
SPAC23C4.17 41.2 7e-04
7300487 40.8 0.001
At5g26180 39.7 0.002
Hs14149837 39.7 0.002
7300488 39.7 0.002
At4g26600 37.7 0.008
ECU08g0400 33.1 0.23
Hs11545785 30.8 1.2
CE25405 30.0 1.7
CE25404_3 30.0 1.7
At3g21465 29.6 2.1
Hs20532638 29.6 2.4
CE24567 28.9 4.4
Hs19923393_2 27.7 8.4
Hs9961357 27.7 9.7
> Hs5453792
Length=855
Score = 74.3 bits (181), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 54/177 (30%), Positives = 77/177 (43%), Gaps = 26/177 (14%)
Query 1 GKIYLHDVRERMLQEAKKRLKRAGITNYTIVEPGNPTLSKLLGAMDWVVCDVPCSGTGAL 60
G I +D L+ L R G+TN I K++G D V+ D PCSGTG +
Sbjct 410 GVILANDANAERLKSVVGNLHRLGVTNTIISHYDGRQFPKVVGGFDRVLLDAPCSGTGVI 469
Query 61 RRNPEMKWKYKDEKLMDFVALQRHIFSSALQY--AKPKTGK-IVYITCSILDEENVHQAK 117
++P +K ++ ++ LQ+ + SA+ A KTG +VY TCSI EEN
Sbjct 470 SKDPAVKTNKDEKDILRCAHLQKELLLSAIDSVNATSKTGGYLVYCTCSITVEENEWVVD 529
Query 118 FFCQKHGLVLT---------------EPPFHS--------LPTSHGMDGFFCAIFSR 151
+ +K + L E FH P +H MDGFF A F +
Sbjct 530 YALKKRNVRLVPTGLDFGQEGFTRFRERRFHPSLRSTRRFYPHTHNMDGFFIAKFKK 586
> Hs20556018
Length=812
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 54/177 (30%), Positives = 77/177 (43%), Gaps = 26/177 (14%)
Query 1 GKIYLHDVRERMLQEAKKRLKRAGITNYTIVEPGNPTLSKLLGAMDWVVCDVPCSGTGAL 60
G I +D L+ L R G+TN I K++G D V+ D PCSGTG +
Sbjct 410 GVILANDANAERLKSVVGNLHRLGVTNTIISHYDGRQFPKVVGGFDRVLLDAPCSGTGVI 469
Query 61 RRNPEMKWKYKDEKLMDFVALQRHIFSSALQY--AKPKTGK-IVYITCSILDEENVHQAK 117
++P +K ++ ++ LQ+ + SA+ A KTG +VY TCSI EEN
Sbjct 470 SKDPAVKTNKDEKDILRCAHLQKELLLSAIDSVNATSKTGGYLVYCTCSITVEENEWVVD 529
Query 118 FFCQKHGLVLT---------------EPPFHS--------LPTSHGMDGFFCAIFSR 151
+ +K + L E FH P +H MDGFF A F +
Sbjct 530 YALKKRNVRLVPTGLDFGQEGFTRFRERRFHPSLRSTRRFYPHTHNMDGFFIAKFKK 586
> At3g13180
Length=515
Score = 67.8 bits (164), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 51/173 (29%), Positives = 74/173 (42%), Gaps = 29/173 (16%)
Query 1 GKIYLHDVRE---RMLQEAKKRLKRAGI-----TNYTIVEPGNPTLSKLLGAMDWVVCDV 52
G IY DV E R+L E K + G+ ++ + N D V+ D
Sbjct 348 GMIYAMDVNEGRLRILGETAKSHQVDGLITTIHSDLRVFAETNEV------QYDKVLLDA 401
Query 53 PCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYITCSILDEEN 112
PCSG G L + +++W K E +++ LQ + SA + K G +VY TCSI EEN
Sbjct 402 PCSGLGVLSKRADLRWNRKLEDMLELTKLQDELLDSASKLVK-HGGVLVYSTCSIDPEEN 460
Query 113 VHQAKFFCQKH--------------GLVLTEPPFHSLPTSHGMDGFFCAIFSR 151
+ + F +H V ++ F S P H +DG F A R
Sbjct 461 EGRVEAFLLRHPEFTIDPVTSFVPSSFVTSQGFFLSNPVKHSLDGAFAARLVR 513
> At5g55920
Length=682
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 51/175 (29%), Positives = 77/175 (44%), Gaps = 28/175 (16%)
Query 1 GKIYLHDVRERMLQEAKKRLKRAGITNYTIVEPGNPTLSKLLG--AMDWVVCDVPCSGTG 58
G IY ++++ L+ L R G+TN + L K+LG +D V+ D PCSGTG
Sbjct 385 GLIYANEMKVPRLKSLTANLHRMGVTNTIVCNYDGRELPKVLGQNTVDRVLLDAPCSGTG 444
Query 59 ALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQY--AKPKTGK-IVYITCSILDEENVHQ 115
+ ++ +K +++ F LQ+ + +A+ A KTG IVY TCSI+ EN
Sbjct 445 IISKDESVKITKTMDEIKKFAHLQKQLLLAAIDMVDANSKTGGYIVYSTCSIMVTENEAV 504
Query 116 AKFFCQKHGLVLT-------------------EPPFHSL----PTSHGMDGFFCA 147
+ +K + L +P P H MDGFF A
Sbjct 505 IDYALKKRDVKLVTCGLDFGRKGFTRFREHRFQPSLDKTRRFYPHVHNMDGFFVA 559
> ECU01g1080
Length=370
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 61/122 (50%), Gaps = 3/122 (2%)
Query 1 GKIYLHDVRERMLQEAKKRLKRAGITNYTIVEPGNPTLSKLLGAMDWVVCDVPCSGTGAL 60
G IY +DV E + K ++R G+ N ++ ++ +G +D V+ D PCSGTG +
Sbjct 197 GVIYANDVNEERIAGLKSNIQRMGVRNCIVMNMDGRKVN--VGRVDRVLLDAPCSGTGVI 254
Query 61 RRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYITCSILDEENVHQAKFFC 120
++P +K ++ V LQ+ + KP G ++Y TCS+L +EN +
Sbjct 255 SKDPSVKTSRTRSEIDRMVTLQKELILHGFGMLKPG-GILMYSTCSVLVKENEEVVNYLL 313
Query 121 QK 122
K
Sbjct 314 SK 315
> YNL061w
Length=618
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 42/178 (23%), Positives = 74/178 (41%), Gaps = 27/178 (15%)
Query 1 GKIYLHDVRERMLQEAKKRLKRAGITNYTIVEPGNPTLSKLLGAMDWVVCDVPCSGTGAL 60
G ++ +D + + + R G TN + K++G D ++ D PCSGTG +
Sbjct 371 GCVFANDANKSRTKSLIANIHRLGCTNTIVCNYDAREFPKVIGGFDRILLDAPCSGTGVI 430
Query 61 RRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKT---GKIVYITCSILDEENVHQAK 117
++ +K ++ + LQ+ + SA+ + G IVY TCS+ EE+
Sbjct 431 GKDQSVKVSRTEKDFIQIPHLQKQLLLSAIDSVDCNSKHGGVIVYSTCSVAVEEDEAVID 490
Query 118 FFCQKH--------GLVLTEPPFHS----------------LPTSHGMDGFFCAIFSR 151
+ +K GL + + F S P ++ +DGFF A F +
Sbjct 491 YALRKRPNVKLVDTGLAIGKEAFTSYRGKKFHPSVKLARRYYPHTYNVDGFFVAKFQK 548
> CE28259
Length=664
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 60/131 (45%), Gaps = 13/131 (9%)
Query 20 LKRAGITNYTIVEPGNPTLSKLL-GAMDWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDF 78
L R G+ N + G SK+ D ++ D PCSGTG + ++ +K + +
Sbjct 350 LHRLGVNNTVVCNLGGEEFSKIRPNGFDRILLDAPCSGTGVIWKDQSVKTSKDSQDVQRR 409
Query 79 VALQRHIFSSALQY---AKPKTGKIVYITCSILDEENVHQAKFFCQKHGLVLTEPPFHSL 135
+QR + SAL P G +VY TCS+L EEN F ++ L +
Sbjct 410 HTMQRQLILSALDSLDANSPNGGYLVYSTCSVLVEENEAVVNFLLERRHCEL-------V 462
Query 136 PT--SHGMDGF 144
PT S G+DG+
Sbjct 463 PTGLSIGVDGY 473
> 7303448
Length=857
Score = 57.8 bits (138), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 45/177 (25%), Positives = 73/177 (41%), Gaps = 26/177 (14%)
Query 1 GKIYLHDVRERMLQEAKKRLKRAGITNYTIVEPGNPTLSKLLGAMDWVVCDVPCSGTGAL 60
G ++ +D ++ R GI N + ++ D V+ D PC+GTG +
Sbjct 427 GVLFANDSNRDRIKAIVANFHRLGIVNAVVSCEDGTKFRNIMTGFDRVLLDAPCTGTGVV 486
Query 61 RRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKT---GKIVYITCSILDEENVHQAK 117
++P +K + + LQR + +A+ K+ G IVY TCS+L EEN
Sbjct 487 SKDPSVKTTKSEVDVQRCYNLQRKLLLTAIDCTDAKSSTGGYIVYSTCSVLPEENEWVID 546
Query 118 FFCQKHGLVLT---------------EPPFH-SL-------PTSHGMDGFFCAIFSR 151
+ +K + L + FH SL P +H MDGF+ A +
Sbjct 547 YALKKRNVKLVPTGLDFGVEGFTKFRQHRFHPSLNLTKRYYPHTHNMDGFYVAKLKK 603
> SPBP8B7.20c
Length=608
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 51/175 (29%), Positives = 76/175 (43%), Gaps = 28/175 (16%)
Query 1 GKIYLHDVRERMLQEAKKRLKRAGITNYTIVE-PGNPTLSKLLGAMDWVVCDVPCSGTGA 59
G I+ +D + + + R G+ N + G ++++G D V+ D PCSGTG
Sbjct 365 GIIFANDSNKARTKALSANIHRLGVRNAIVCNYDGRKFPNEVIGGFDRVLLDAPCSGTGV 424
Query 60 LRRNPEMKWKYKDEKLMDFVALQRHIFSSALQY--AKPKTGK-IVYITCSILDEENVHQA 116
+ ++ +K + LQR + SA+ A KTG IVY TCSI +E+
Sbjct 425 IYKDQSVKTNKSERDFDTLSHLQRQLLLSAIDSVNADSKTGGFIVYSTCSITVDEDEAVI 484
Query 117 KFFCQKH---GLVLT-------------EPPFH-SL-------PTSHGMDGFFCA 147
++ +K LV T E FH SL P H +DGFF A
Sbjct 485 QYALKKRPNVKLVSTGLEFGREGFTRFREKRFHPSLKLTRRYYPHVHNIDGFFVA 539
> Hs20473687
Length=469
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 47/87 (54%), Gaps = 4/87 (4%)
Query 44 AMDWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYI 103
+ D ++ D PCSG G + P M + +++ + LQR +F++A+Q KP+ G +VY
Sbjct 316 SFDRILLDAPCSGMG---QRPNMACTWSVKEVASYQPLQRKLFTAAVQLLKPE-GVLVYS 371
Query 104 TCSILDEENVHQAKFFCQKHGLVLTEP 130
TC+I EN Q + K + +P
Sbjct 372 TCTITLAENEEQVAWALTKFPCLQLQP 398
> 7290467
Length=746
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 42/77 (54%), Gaps = 1/77 (1%)
Query 36 PTLSKLLGAMDWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKP 95
P SK + D ++CDVPCSG G LR+NP++ K+ + + +Q I + +
Sbjct 255 PDGSKAILKFDKILCDVPCSGDGTLRKNPDIWLKWNLAQAYNLHGIQYRIVRRGAEMLE- 313
Query 96 KTGKIVYITCSILDEEN 112
G++VY TCS+ EN
Sbjct 314 VGGRLVYSTCSLNPIEN 330
> YBL024w
Length=684
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 52/112 (46%), Gaps = 9/112 (8%)
Query 9 RERMLQEAKKRLKRAG--ITNYT------IVEPGNPTLSKLLGAMDWVVCDVPCSGTGAL 60
R ML KRL A + N+ I GN + D ++CDVPCSG G +
Sbjct 207 RSHMLVHQLKRLNSANLMVVNHDAQFFPRIRLHGNSNNKNDVLKFDRILCDVPCSGDGTM 266
Query 61 RRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYITCSILDEEN 112
R+N + + + + A+Q +I + L K G++VY TCS+ EN
Sbjct 267 RKNVNVWKDWNTQAGLGLHAVQLNILNRGLHLLK-NNGRLVYSTCSLNPIEN 317
> Hs8922322
Length=429
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 32/121 (26%), Positives = 52/121 (42%), Gaps = 15/121 (12%)
Query 1 GKIYLHDVRERMLQEAKKRLKRAGIT-------NYTIVEPGNPTLSKLLGAMDWVVCDVP 53
GKI+ D+ + L L RAG++ ++ V P +P + + +++ D
Sbjct 252 GKIFAFDLDAKRLASMATLLARAGVSCCELAEEDFLAVSPSDPRYHE----VHYILLDPS 307
Query 54 CSGTGALRRNPEMKWKYKDE--KLMDFVALQRHIFSSALQYAKPKTGKIVYITCSILDEE 111
CSG+G R E +L Q+ AL + P ++VY TCS+ EE
Sbjct 308 CSGSGMPSRQLEEPGAGTPSPVRLHALAGFQQRALCHALTF--PSLQRLVYSTCSLCQEE 365
Query 112 N 112
N
Sbjct 366 N 366
> At2g22400
Length=837
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 35/64 (54%), Gaps = 1/64 (1%)
Query 44 AMDWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYI 103
A D V+CDVPCSG G LR+ P++ K+ +LQ + L K GK++Y
Sbjct 281 AFDRVLCDVPCSGDGTLRKAPDIWRKWNSGMGNGLHSLQIILAMRGLSLLK-VGGKMIYS 339
Query 104 TCSI 107
TCS+
Sbjct 340 TCSM 343
> Hs8923285
Length=531
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 37/63 (58%), Gaps = 3/63 (4%)
Query 46 DWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFS-SALQYAKPKTGKIVYIT 104
D ++CDVPCSG G +R+N ++ K+ + LQ I + A Q A + G++VY T
Sbjct 27 DRILCDVPCSGDGTMRKNIDVWKKWTTLNSLQLHGLQLRIATRGAEQLA--EGGRVVYST 84
Query 105 CSI 107
CS+
Sbjct 85 CSL 87
> 7296593
Length=358
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 39/69 (56%), Gaps = 5/69 (7%)
Query 44 AMDWVVCDVPCSGTGALRRNPEMKWKYKDEKLM-DFVALQRHIFSSALQYAKPKTGKIVY 102
+ D ++ D PCSG G P++ K K++ + +QR +F A+Q +P G +VY
Sbjct 229 SFDRILLDAPCSGLG---NRPQLSCSIKQVKVLSSYPHIQRRLFEQAVQLVRPG-GILVY 284
Query 103 ITCSILDEE 111
TC++ ++E
Sbjct 285 STCTVTEDE 293
> SPAC17D4.04
Length=396
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 36/67 (53%), Gaps = 1/67 (1%)
Query 46 DWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYITC 105
D ++ DVPCSG G R+N + ++ + + A Q I LQ + K G++VY TC
Sbjct 235 DRILADVPCSGDGTFRKNIALWNEWSLKTALGLHATQIKILMRGLQLLE-KGGRLVYSTC 293
Query 106 SILDEEN 112
S+ EN
Sbjct 294 SLNPIEN 300
> CE22185
Length=203
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 38/73 (52%), Gaps = 4/73 (5%)
Query 46 DWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYITC 105
D V+ DV CSG G LR+NPE+ K+ + + +Q I Q K G++VY TC
Sbjct 52 DRVLADVICSGDGTLRKNPEIWKKWTPQDGLGLHRMQIAIARKGAQQLKV-GGRMVYSTC 110
Query 106 S---ILDEENVHQ 115
S I DE V Q
Sbjct 111 SMNPIEDEAVVAQ 123
> At4g40000
Length=682
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 34/62 (54%), Gaps = 1/62 (1%)
Query 46 DWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYITC 105
D V+CDVPCSG G LR+ P++ ++ +LQ + L K G++VY TC
Sbjct 174 DRVLCDVPCSGDGTLRKAPDIWRRWNSGSGNGLHSLQVVLAMRGLSLLKV-GGRMVYSTC 232
Query 106 SI 107
S+
Sbjct 233 SM 234
> ECU07g0320
Length=364
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 43/174 (24%), Positives = 75/174 (43%), Gaps = 36/174 (20%)
Query 1 GKIYLHDVRERMLQEAKKRLKRAGITNYTIVEP----GNPTLSKLLGAMDWVVCDVPCSG 56
GKIY + + + + +L + G++N +VE NP + + +++CD CSG
Sbjct 202 GKIYAFERSKNRAETLRAQLFKLGVSNTEVVEDDFMNANP---EDFEGIRYILCDPSCSG 258
Query 57 TGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYITCS---------- 106
+G + +K E++ + Q I AL++ +P+ K+VY CS
Sbjct 259 SGM-----HLGYKMDKERIEGLKSFQIEIVRHALRF-RPE--KLVYSVCSDHREEGEEVV 310
Query 107 --IL-----DEENVHQAKFFCQKH--GLVLTEPPFHSLPTSHGMDGFFCAIFSR 151
IL + EN+ + F+ H G ++ G GFF A+F R
Sbjct 311 EEILRSSEYELENI--SMFWSSAHSSGFSFSQDVVRCRRDESGAIGFFIALFVR 362
> SPAC23C4.17
Length=674
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query 46 DWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYITC 105
D ++ DVPCSG G R+N + ++ LQ I LQ K G +VY TC
Sbjct 265 DRILADVPCSGDGTFRKNLSLWREWSANSAFSLHPLQLRILIRGLQLLK-VGGCLVYSTC 323
Query 106 SILDEEN 112
SI EN
Sbjct 324 SINPIEN 330
> 7300487
Length=433
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 35/68 (51%), Gaps = 1/68 (1%)
Query 45 MDWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYIT 104
+++++ D CSG+G R +D++L LQ I S A+ A P +I Y T
Sbjct 298 VEYILVDPSCSGSGMQNRMTVCDEPKEDKRLQKLQGLQIKILSHAMG-AFPNVKRIAYCT 356
Query 105 CSILDEEN 112
CS+ EEN
Sbjct 357 CSLWKEEN 364
> At5g26180
Length=532
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 44/183 (24%), Positives = 71/183 (38%), Gaps = 38/183 (20%)
Query 1 GKIYLHDVRERMLQEAKKRLKRAGIT----NYTIVEPGNPTLSKLLGAMDWVVCDVPCSG 56
GKI ++ E ++ + +K + I ++ + P +P+ +K+ ++ D CSG
Sbjct 330 GKIIACELNEERVKRLEHTIKLSDIEVCHGDFLGLNPKDPSFAKIRA----ILLDPSCSG 385
Query 57 TGAL---------RRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYITCSI 107
+G + + + Y +L Q+ + AL + PK ++VY TCSI
Sbjct 386 SGTITDRLDHLLPSHSEDNNMNYDSMRLHKLAVFQKKALAHALSF--PKVERVVYSTCSI 443
Query 108 LDEEN---VHQAKFFCQKHGLVLTEP----PFHSLPTSHG------MD------GFFCAI 148
EN V G L P LP G MD GFF A+
Sbjct 444 YQIENEDVVSSVLPLASSLGFKLATPFPQWQRRGLPVFAGSEHLLRMDPVEDKEGFFIAL 503
Query 149 FSR 151
F R
Sbjct 504 FVR 506
> Hs14149837
Length=315
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/121 (26%), Positives = 50/121 (41%), Gaps = 15/121 (12%)
Query 1 GKIYLHDVRERMLQEAKKRLKRAGIT-------NYTIVEPGNPTLSKLLGAMDWVVCDVP 53
GKI+ D+ R L L AG++ ++ V P +P ++ +V+ D
Sbjct 68 GKIFAFDLDARRLASMATLLAWAGVSCCELAEEDFLAVSPLDPRYREV----HYVLLDPS 123
Query 54 CSGTGALRRNPEMKWKYKDE--KLMDFVALQRHIFSSALQYAKPKTGKIVYITCSILDEE 111
CSG+G R E +L Q+ AL + P ++VY CS+ EE
Sbjct 124 CSGSGMPSRQLEEPGAGTPSPVRLHALAGFQQRALCHALTF--PSLQRLVYSMCSLCQEE 181
Query 112 N 112
N
Sbjct 182 N 182
> 7300488
Length=383
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 35/68 (51%), Gaps = 1/68 (1%)
Query 45 MDWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYIT 104
+++++ D CSG+G R +D++L LQ I S A+ A P +I Y T
Sbjct 248 VEYILVDPSCSGSGMQNRMTVCDEPKEDKRLQKLQGLQIKILSHAMG-AFPNVKRIAYCT 306
Query 105 CSILDEEN 112
CS+ EEN
Sbjct 307 CSLWKEEN 314
> At4g26600
Length=626
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 33/133 (24%), Positives = 51/133 (38%), Gaps = 25/133 (18%)
Query 42 LGAMDWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQY--AKPKTGK 99
+G + +VC+ G + ++ +K + + F LQ+ + A+ A KTG
Sbjct 390 MGVTNTIVCNY--DGREVISKDESVKTSKSADDIKKFAHLQKQLILGAIDLVDANSKTGG 447
Query 100 -IVYITCSILDEENVHQAKFFCQKH-------GLVLTEPPFHS-------------LPTS 138
IVY TCS++ EN + + GL P F P
Sbjct 448 YIVYSTCSVMIPENEAVIDYALKNRDVKLVPCGLDFGRPGFREHRFHPSLEKTRRFYPHV 507
Query 139 HGMDGFFCAIFSR 151
H MDGFF A +
Sbjct 508 HNMDGFFVAKLKK 520
> ECU08g0400
Length=515
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 30/67 (44%), Gaps = 1/67 (1%)
Query 46 DWVVCDVPCSGTGALRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYITC 105
D V CDV CS G +R++P + ++K K Q I + G + Y TC
Sbjct 188 DRVFCDVICSSDGTMRKSPGILDEWKLSKSGGLFDTQWRILKRGCDLV-AEGGLVSYSTC 246
Query 106 SILDEEN 112
S+ EN
Sbjct 247 SLNPIEN 253
> Hs11545785
Length=340
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 37/83 (44%), Gaps = 2/83 (2%)
Query 46 DWVVCDVPCSGTGA-LRRNPEMKWKYKDEKLMDFVALQRHIFSSALQYAKPKTGKIVYIT 104
D V+ D PCS + L + K + + + LQ + SA++ +P G +VY T
Sbjct 206 DKVLVDAPCSNDRSWLFSSDSQKASCRISQRRNLPLLQIELLRSAIKALRP-GGILVYST 264
Query 105 CSILDEENVHQAKFFCQKHGLVL 127
C++ EN HG ++
Sbjct 265 CTLSKAENQDVISEILNSHGNIM 287
> CE25405
Length=469
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 39/165 (23%), Positives = 58/165 (35%), Gaps = 55/165 (33%)
Query 40 KLLGAMDWVVCDVPCSGTGALRRNPEMK-WKYKDEKLMDFVALQRH-------------- 84
K + + + D PCSG+G ++R E+ + E+L LQ H
Sbjct 302 KKFSKVKFAIVDPPCSGSGIVKRMDEITGGNAEKERLEKLKNLQVHKNLDLLKMCWKCSE 361
Query 85 ----------------IFSSALQYAKPKTGKIVYITCSILDEENVH-----------QAK 117
I AL+ P + VY TCS+ +EEN +
Sbjct 362 NCFKKKTKKLIENDAMILKHALKL--PGLKRAVYSTCSVHEEENEQVVDEVLLDTYVRQN 419
Query 118 FFCQKH--------GLVLTEPPFHSL---PTSHGMDGFFCAIFSR 151
+ +K+ GL E H L P +GFF A+F R
Sbjct 420 YVLKKNVLPEWTYRGLKTYEVGEHCLRANPKVTLTNGFFVAVFER 464
> CE25404_3
Length=436
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 39/166 (23%), Positives = 58/166 (34%), Gaps = 55/166 (33%)
Query 39 SKLLGAMDWVVCDVPCSGTGALRRNPEMK-WKYKDEKLMDFVALQRH------------- 84
K + + + D PCSG+G ++R E+ + E+L LQ H
Sbjct 268 DKKFSKVKFAIVDPPCSGSGIVKRMDEITGGNAEKERLEKLKNLQVHKNLDLLKMCWKCS 327
Query 85 -----------------IFSSALQYAKPKTGKIVYITCSILDEENVH-----------QA 116
I AL+ P + VY TCS+ +EEN +
Sbjct 328 ENCFKKKTKKLIENDAMILKHALKL--PGLKRAVYSTCSVHEEENEQVVDEVLLDTYVRQ 385
Query 117 KFFCQKH--------GLVLTEPPFHSL---PTSHGMDGFFCAIFSR 151
+ +K+ GL E H L P +GFF A+F R
Sbjct 386 NYVLKKNVLPEWTYRGLKTYEVGEHCLRANPKVTLTNGFFVAVFER 431
> At3g21465
Length=246
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 35/157 (22%), Positives = 56/157 (35%), Gaps = 30/157 (19%)
Query 13 LQEAKKRLKRAGITNYTIVEPGNPTLSKLLGAMDWVVCDVPCSGTGALRRNPEMKWKYKD 72
L + + L+R ++N I + N L + + C GA+ WK+
Sbjct 76 LFDVLQNLRRFRLSNLRIHDNFNCNLCQQVAK--------TCVRVGAINHGKRALWKHNV 127
Query 73 EKLMDFVALQRHIFSSALQYA-------------------KPKTGKIVYITCSILDEEN- 112
L VA H+ S AL++ +P T +V+ C D +
Sbjct 128 HGLTPSVASAHHLLSYALKHKDAKLMDEVMKLLKMNNLPLQPGTADLVFRICHDTDNWDL 187
Query 113 -VHQAKFFCQKHGLVLTEPPFHSLPTSHGMDGFFCAI 148
V +K FC K G+ L + F G FC +
Sbjct 188 LVKYSKKFC-KAGVKLRKTTFDVWMEFAAKRGTFCVV 223
> Hs20532638
Length=454
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 28/49 (57%), Gaps = 3/49 (6%)
Query 13 LQEAKKRLKRAGI--TNYTIVEPGNPTLSKLLGAMDWVVCDVPCSGTGA 59
L++A+ + G+ N +++ G ++ L GAM WV C++ +G GA
Sbjct 40 LEKAQHSQSKTGLRCGNESVMN-GQASVHALFGAMAWVSCNLVTTGKGA 87
> CE24567
Length=447
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 36/82 (43%), Gaps = 21/82 (25%)
Query 54 CSGTGALRRNPEMKWKYKDEKL-----------MDFVALQRHIFSSALQYAKPKTGKIVY 102
CSGTG++R +K +K+EK + F A + IF Y P
Sbjct 273 CSGTGSVRNYTRIKVYFKNEKSEHYTESEIPEKLLFQAEGKRIFEEEQDYIIP------- 325
Query 103 ITCSILDEENVHQ-AKFFCQKH 123
S EE+V++ +K FC +H
Sbjct 326 --ISQYQEEDVNKMSKLFCAQH 345
> Hs19923393_2
Length=1458
Score = 27.7 bits (60), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 102 YITCSILDEENVHQAKFFCQKHGLVLTEPPFHSLPTS 138
Y CSI D + VH ++ C+ L +PP SLP +
Sbjct 14 YFICSIQDFKLVHNSQACCRSPTPALCDPPACSLPVA 50
> Hs9961357
Length=661
Score = 27.7 bits (60), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 23/45 (51%), Gaps = 6/45 (13%)
Query 49 VCDVPCSGTGAL--RRNPEMKWKYKDE----KLMDFVALQRHIFS 87
VC PC+ R N EMKWKY+ + L+DFV Q + S
Sbjct 449 VCLEPCTVNVDYMNRNNIEMKWKYEGKVLHGDLIDFVCKQGYDLS 493
Lambda K H
0.323 0.138 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1925034518
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40