bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1724_orf2
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
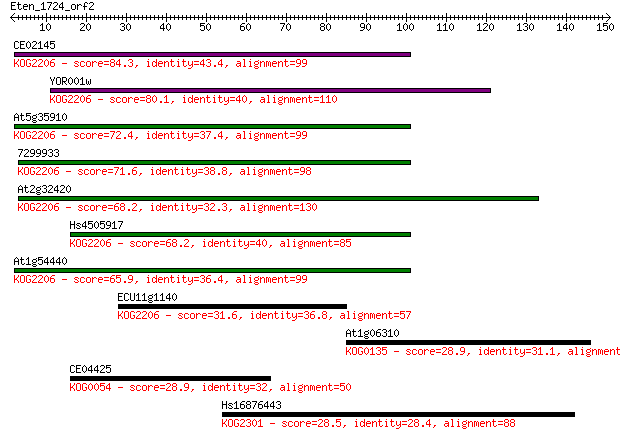
CE02145 84.3 8e-17
YOR001w 80.1 1e-15
At5g35910 72.4 3e-13
7299933 71.6 5e-13
At2g32420 68.2 5e-12
Hs4505917 68.2 5e-12
At1g54440 65.9 3e-11
ECU11g1140 31.6 0.63
At1g06310 28.9 3.5
CE04425 28.9 4.0
Hs16876443 28.5 5.5
> CE02145
Length=876
Score = 84.3 bits (207), Expect = 8e-17, Method: Composition-based stats.
Identities = 43/99 (43%), Positives = 60/99 (60%), Gaps = 7/99 (7%)
Query 2 PFMTVYRKQQLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSL 61
P + K++L + L S K A+D EHH SY G CLIQ+ST +D +
Sbjct 277 PLTMIDTKEKLEALTKTLNS--VKEFAVDLEHHQMRSYLGLTCLIQIST-----RDEDFI 329
Query 62 VDPFSLFVHLTALNELTANPKILKILHGSASDVIWLRRE 100
+DPF ++ H+ LNE ANP+ILK+ HGS SDV+WL+R+
Sbjct 330 IDPFPIWDHVGMLNEPFANPRILKVFHGSDSDVLWLQRD 368
> YOR001w
Length=733
Score = 80.1 bits (196), Expect = 1e-15, Method: Composition-based stats.
Identities = 44/110 (40%), Positives = 63/110 (57%), Gaps = 8/110 (7%)
Query 11 QLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPFSLFVH 70
+L ML++L K +A+D EHH Y SY G +CL+Q+ST +D LVD L +
Sbjct 221 ELESMLEDL--KNTKEIAVDLEHHDYRSYYGIVCLMQIST-----RERDYLVDTLKLREN 273
Query 71 LTALNELTANPKILKILHGSASDVIWLRRESLGVMTTPSEPTSHQGRPDG 120
L LNE+ NP I+K+ HG+ D+IWL+R+ LG+ T H + G
Sbjct 274 LHILNEVFTNPSIVKVFHGAFMDIIWLQRD-LGLYVVGLFDTYHASKAIG 322
> At5g35910
Length=838
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 60/100 (60%), Gaps = 8/100 (8%)
Query 2 PFMTVYRKQQLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSL 61
PF V + L+E++ +L S A+D EH+ Y S++G CL+Q+ST +D +
Sbjct 227 PFKFVQEVKDLKELVAKLRSVEE--FAVDLEHNQYRSFQGLTCLMQIST-----RTEDYI 279
Query 62 VDPFSLFVHL-TALNELTANPKILKILHGSASDVIWLRRE 100
VD F L +H+ L E+ +PK K++HG+ D+IWL+R+
Sbjct 280 VDTFKLRIHIGPYLREIFKDPKKKKVMHGADRDIIWLQRD 319
> 7299933
Length=900
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 38/98 (38%), Positives = 57/98 (58%), Gaps = 7/98 (7%)
Query 3 FMTVYRKQQLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLV 62
M V ++L++ L+EL +AID EHHSY ++ G CL+Q+ST +KD +
Sbjct 271 LMVVDTVEKLKQALEELRQAPQ--IAIDVEHHSYRTFMGITCLVQMST-----RSKDYIF 323
Query 63 DPFSLFVHLTALNELTANPKILKILHGSASDVIWLRRE 100
D L + LN + +PK LKILHG+ D+ WL+R+
Sbjct 324 DTLILRDDMHILNLVLTDPKKLKILHGADLDIEWLQRD 361
> At2g32420
Length=237
Score = 68.2 bits (165), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 42/133 (31%), Positives = 71/133 (53%), Gaps = 10/133 (7%)
Query 3 FMTVYRKQQLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLV 62
++ V + QL+E+ + L + +V A+DTE HS S+ GF LIQ+ST +D LV
Sbjct 55 YVWVETESQLKELAEIL--AKEQVFAVDTEQHSLRSFLGFTALIQIST-----HEEDFLV 107
Query 63 DPFSLFVHLTALNELTANPKILKILHGSASDVIWLRRE---SLGVMTTPSEPTSHQGRPD 119
D +L ++ L + ++P I K+ HG+ +DVIWL+R+ + M ++ +P
Sbjct 108 DTIALHDVMSILRPVFSDPNICKVFHGADNDVIWLQRDFHIYVVNMFDTAKACEVLSKPQ 167
Query 120 GNVQSELHNACGA 132
++ L CG
Sbjct 168 RSLAYLLETVCGV 180
> Hs4505917
Length=860
Score = 68.2 bits (165), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 52/90 (57%), Gaps = 10/90 (11%)
Query 16 LDELCSGRHKVV-----AIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPFSLFVH 70
LDEL K++ A+D EHHSY S+ G CL+Q+ST +D ++D L
Sbjct 294 LDELVELNEKLLNCQEFAVDLEHHSYRSFLGLTCLMQIST-----RTEDFIIDTLELRSD 348
Query 71 LTALNELTANPKILKILHGSASDVIWLRRE 100
+ LNE +P I+K+ HG+ SD+ WL+++
Sbjct 349 MYILNESLTDPAIVKVFHGADSDIEWLQKD 378
> At1g54440
Length=738
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 36/100 (36%), Positives = 57/100 (57%), Gaps = 8/100 (8%)
Query 2 PFMTVYRKQQLREMLDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSL 61
PF V + L ++ L S A+D EH+ Y +++G CL+Q+ST +D +
Sbjct 227 PFKLVEEVKDLEDLAAALQSVEE--FAVDLEHNQYRTFQGLTCLMQIST-----RTEDYI 279
Query 62 VDPFSLFVHLTA-LNELTANPKILKILHGSASDVIWLRRE 100
VD F L+ H+ L EL +PK K++HG+ D+IWL+R+
Sbjct 280 VDIFKLWDHIGPYLRELFKDPKKKKVIHGADRDIIWLQRD 319
> ECU11g1140
Length=359
Score = 31.6 bits (70), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 31/60 (51%), Gaps = 9/60 (15%)
Query 28 AIDTEHHSYESYKGFICLIQLSTCGSAGAAKDS--LVDPFSLFVHLTALNELTAN-PKIL 84
AI+ H+Y SY+GF+C I SAG + + ++D L + L L N PKI+
Sbjct 135 AIEVFDHTYRSYEGFVCFI------SAGDTRGNVYVIDAIKLRTVIPRLRLLGCNVPKIV 188
> At1g06310
Length=667
Score = 28.9 bits (63), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 31/66 (46%), Gaps = 5/66 (7%)
Query 85 KILHGSASDVIWLRRESLGVMTTPSEPT----SHQGRPD-GNVQSELHNACGARHPHRIR 139
K+ GS DV+ L R +++ +P+ H R + G+V+ E+ CG PH +
Sbjct 580 KLPPGSVKDVLGLVRSMYALISLEEDPSLLRYGHLSRDNVGDVRKEVSKLCGELRPHALA 639
Query 140 QGCSAG 145
S G
Sbjct 640 LVASFG 645
> CE04425
Length=1421
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Query 16 LDELCSGRHKVVAIDTEHHSYESYKGFICLIQLSTCGSAGAAKDSLVDPF 65
L E CSGR + +D +H S +G + +I +G +++L DPF
Sbjct 1184 LKEYCSGRVTIDGVDLDHISLNFRRGGMSIIPQEPVIFSGTVRENL-DPF 1232
> Hs16876443
Length=528
Score = 28.5 bits (62), Expect = 5.5, Method: Composition-based stats.
Identities = 25/92 (27%), Positives = 36/92 (39%), Gaps = 8/92 (8%)
Query 54 AGAAKDSLVDPFSLFVHLTALNELTANPKILKILHGSASDVIWLRRE----SLGVMTTPS 109
A A + L+D FSL HL L++ I ++L S + L + +
Sbjct 15 ADAIRSRLIDTFSLIEHLQGLSQAVPRHTIRELLDPSRQKKLVLGDQHQLVRFSIKPQRI 74
Query 110 EPTSHQGRPDGNVQSELHNACGARHPHRIRQG 141
E SH R + S LH C R P + G
Sbjct 75 EQISHAQR----LLSRLHVRCSQRPPLSLWAG 102
Lambda K H
0.319 0.134 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1852391706
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40