bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1722_orf1
Length=175
Score E
Sequences producing significant alignments: (Bits) Value
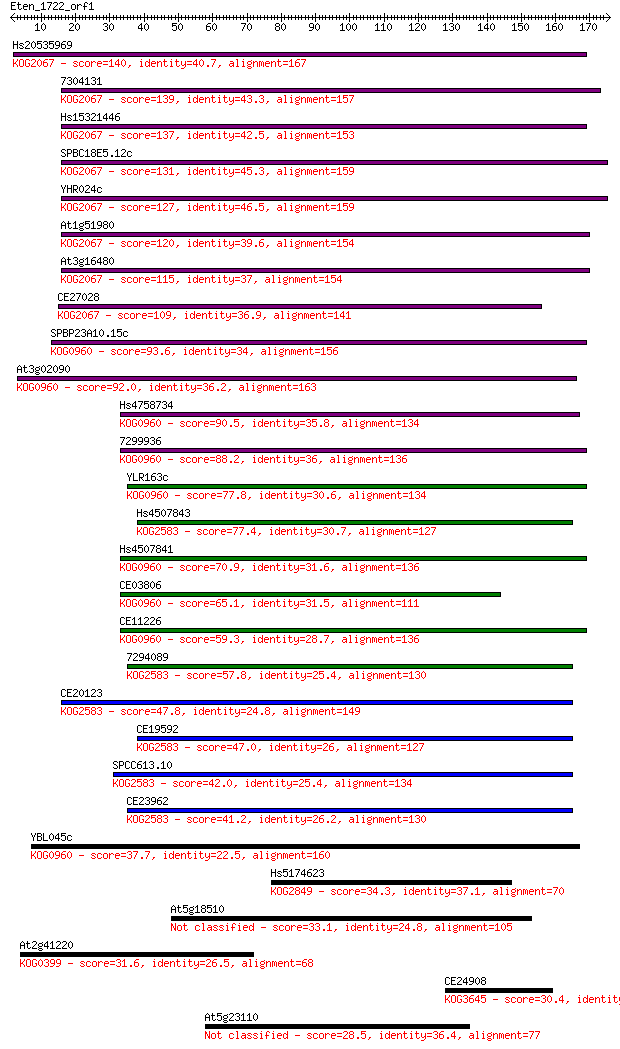
Hs20535969 140 1e-33
7304131 139 3e-33
Hs15321446 137 1e-32
SPBC18E5.12c 131 9e-31
YHR024c 127 1e-29
At1g51980 120 1e-27
At3g16480 115 3e-26
CE27028 109 3e-24
SPBP23A10.15c 93.6 2e-19
At3g02090 92.0 5e-19
Hs4758734 90.5 2e-18
7299936 88.2 8e-18
YLR163c 77.8 1e-14
Hs4507843 77.4 1e-14
Hs4507841 70.9 1e-12
CE03806 65.1 7e-11
CE11226 59.3 4e-09
7294089 57.8 1e-08
CE20123 47.8 1e-05
CE19592 47.0 2e-05
SPCC613.10 42.0 7e-04
CE23962 41.2 0.001
YBL045c 37.7 0.012
Hs5174623 34.3 0.14
At5g18510 33.1 0.31
At2g41220 31.6 0.87
CE24908 30.4 1.9
At5g23110 28.5 6.7
> Hs20535969
Length=417
Score = 140 bits (353), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 68/168 (40%), Positives = 103/168 (61%), Gaps = 1/168 (0%)
Query 2 LMGGGGAFSTGGPGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKAS 61
+MGGGG+FS G P KG+++RLYL+VL+++ W+ +A ++ Y DTGL + A+P +
Sbjct 235 MMGGGGSFSAGRPSKGIFSRLYLDVLSRHHWIYNATSYYLSYEDTGLLCILASADPRQVP 294
Query 62 SAVKVMAEQFAGMT-RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISP 120
V++ ++F M V A EL+RAK + S + MNLE+R ++ ED+GRQ+L + P
Sbjct 295 EMVEITTKEFILMGGTVDAVELERAKTQLTSMLMMNLESRPVIFEDVGRQVLATRSRKLP 354
Query 121 AEFCRAIDNVKEADIRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAAL 168
E C I N K D++RVA K+ P V A G +P Y+ I+ AL
Sbjct 355 QELCTFIRNEKPEDVKRVASKILQGKPVVAALGKSTDLPAYKHIQTAL 402
> 7304131
Length=556
Score = 139 bits (350), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 68/158 (43%), Positives = 101/158 (63%), Gaps = 1/158 (0%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
KGMY+RLY VLN+Y W+ SA A+N Y D GLF ++ A P + V+V+ + GM
Sbjct 387 KGMYSRLYTKVLNRYHWMYSATAYNHAYGDCGLFCVHGSAPPQHMNDMVEVLTREMMGMA 446
Query 76 RVTA-EELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEAD 134
EEL R+K ++S + MNLE+R +V ED+GRQ+L++G+ P F + I++V AD
Sbjct 447 AEPGREELMRSKIQLQSMLLMNLESRPVVFEDVGRQVLVTGQRKRPQHFIKEIESVTAAD 506
Query 135 IRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALRAAG 172
I+RVA+++ S PP+V A GDI ++P I A+ +G
Sbjct 507 IQRVAQRLLSSPPSVAARGDIHNLPEMSHITNAVSGSG 544
> Hs15321446
Length=525
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 65/154 (42%), Positives = 97/154 (62%), Gaps = 1/154 (0%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
KGM++RLYLNVLN++ W+ +A +++ Y DTGL ++ A+P + V+++ ++F M
Sbjct 357 KGMFSRLYLNVLNRHHWMYNATSYHHSYEDTGLLCIHASADPRQVREMVEIITKEFILMG 416
Query 76 -RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEAD 134
V EL+RAK + S + MNLE+R ++ ED+GRQ+L + P E C I NVK D
Sbjct 417 GTVDTVELERAKTQLTSMLMMNLESRPVIFEDVGRQVLATRSRKLPHELCTLIRNVKPED 476
Query 135 IRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAAL 168
++RVA KM P V A GD+ +P YE I+ AL
Sbjct 477 VKRVASKMLRGKPAVAALGDLTDLPTYEHIQTAL 510
> SPBC18E5.12c
Length=494
Score = 131 bits (329), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 72/171 (42%), Positives = 107/171 (62%), Gaps = 15/171 (8%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAG-M 74
KGMY+RLYLNVLN+Y WVE+ MAFN YTD+GLFG+++ + A A ++ + +
Sbjct 326 KGMYSRLYLNVLNQYPWVETCMAFNHSYTDSGLFGMFVTILDDAAHLAAPLIIRELCNTV 385
Query 75 TRVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQL-LMSGRVISPAEFCRAIDNVKEA 133
VT+EE +RAKN +KSS+ MNLE+R I +ED+GRQ+ +G I+P E ID + +
Sbjct 386 LSVTSEETERAKNQLKSSLLMNLESRMISLEDLGRQIQTQNGLYITPKEMIEKIDALTPS 445
Query 134 DIRRVAEKMF----SKP------PTVVAYGDICSVPHYEEIRAALRAAGVG 174
D+ RVA ++ S P PTV+ +G++ V ++ A + AG+G
Sbjct 446 DLSRVARRVLTGNVSNPGNGTGKPTVLIHGNVDEV---GDVFALCKKAGIG 493
> YHR024c
Length=482
Score = 127 bits (318), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 74/174 (42%), Positives = 109/174 (62%), Gaps = 18/174 (10%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQ----F 71
KGMY+RLY +VLN+Y +VE+ +AFN Y+D+G+FG+ + P A AV+V+A+Q F
Sbjct 296 KGMYSRLYTHVLNQYYFVENCVAFNHSYSDSGIFGISLSCIPQAAPQAVEVIAQQMYNTF 355
Query 72 AGMT-RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNV 130
A R+T +E+ RAKN +KSS+ MNLE++ + +ED+GRQ+LM GR I E I+++
Sbjct 356 ANKDLRLTEDEVSRAKNQLKSSLLMNLESKLVELEDMGRQVLMHGRKIPVNEMISKIEDL 415
Query 131 KEADIRRVAEKMFSKP----------PTVVAYGDICSVPHYEEIRAALRAAGVG 174
K DI RVAE +F+ TVV GD S + ++ L+A G+G
Sbjct 416 KPDDISRVAEMIFTGNVNNAGNGKGRATVVMQGDRGS---FGDVENVLKAYGLG 466
> At1g51980
Length=503
Score = 120 bits (302), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 61/156 (39%), Positives = 96/156 (61%), Gaps = 2/156 (1%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
KGM++ LY VLN+Y V+S AF + + DTGLFG+Y ++P A+ A+++ A++ +
Sbjct 348 KGMHSWLYRRVLNEYQEVQSCTAFTSIFNDTGLFGIYGCSSPQFAAKAIELAAKELKDVA 407
Query 76 --RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEA 133
+V L RAK + KS++ MNLE+R I EDIGRQ+L G +F +++D +
Sbjct 408 GGKVNQAHLDRAKAATKSAVLMNLESRMIAAEDIGRQILTYGERKPVDQFLKSVDQLTLK 467
Query 134 DIRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALR 169
DI K+ SKP T+ ++GD+ +VP Y+ I + R
Sbjct 468 DIADFTSKVISKPLTMGSFGDVLAVPSYDTISSKFR 503
> At3g16480
Length=448
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 57/156 (36%), Positives = 92/156 (58%), Gaps = 2/156 (1%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
KGM++ LYL +LN++ +S AF + + +TGLFG+Y +P AS ++++A + +
Sbjct 293 KGMHSWLYLRLLNQHQQFQSCTAFTSVFNNTGLFGIYGCTSPEFASQGIELVASEMNAVA 352
Query 76 --RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEA 133
+V + L RAK + KS+I MNLE+R I EDIGRQ+L G +F + +D +
Sbjct 353 DGKVNQKHLDRAKAATKSAILMNLESRMIAAEDIGRQILTYGERKPVDQFLKTVDQLTLK 412
Query 134 DIRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALR 169
DI K+ +KP T+ +GD+ +VP Y+ + R
Sbjct 413 DIADFTSKVITKPLTMATFGDVLNVPSYDSVSKRFR 448
> CE27028
Length=477
Score = 109 bits (272), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 52/142 (36%), Positives = 86/142 (60%), Gaps = 1/142 (0%)
Query 15 GKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM 74
GKGMY R+Y ++N++ W+ SA+A N Y+D+G+F + + P + A+ ++ Q +
Sbjct 313 GKGMYARMYTELMNRHHWIYSAIAHNHSYSDSGVFTVTASSPPENINDALILLVHQILQL 372
Query 75 TR-VTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEA 133
+ V EL RA+ ++S + MNLE R ++ ED+ RQ+L G P E+ I+ V +
Sbjct 373 QQGVEPTELARARTQLRSHLMMNLEVRPVLFEDMVRQVLGHGDRKQPEEYAEKIEKVTNS 432
Query 134 DIRRVAEKMFSKPPTVVAYGDI 155
DI RV E++ + P++V YGDI
Sbjct 433 DIIRVTERLLASKPSLVGYGDI 454
> SPBP23A10.15c
Length=457
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 53/158 (33%), Positives = 91/158 (57%), Gaps = 3/158 (1%)
Query 13 GPGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLA-NPNKASSAVKVMAEQF 71
G + +RL ++ ++ S M+F+T Y+DTGL+G+Y++ N + V + +
Sbjct 296 GASPHLSSRLS-TIVQQHQLANSFMSFSTSYSDTGLWGIYLVTENLGRIDDLVHFTLQNW 354
Query 72 AGMTRVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVK 131
A +T T E++RAK +++S+ ++L++ + EDIGRQLL +GR +SP E I +
Sbjct 355 ARLTVATRAEVERAKAQLRASLLLSLDSTTAIAEDIGRQLLTTGRRMSPQEVDLRIGQIT 414
Query 132 EADIRRVAEKM-FSKPPTVVAYGDICSVPHYEEIRAAL 168
E D+ RVA +M + K V A G I + Y IR+++
Sbjct 415 EKDVARVASEMIWDKDIAVSAVGSIEGLLDYNRIRSSI 452
> At3g02090
Length=531
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 59/165 (35%), Positives = 88/165 (53%), Gaps = 12/165 (7%)
Query 3 MGGGGAFSTGGPGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASS 62
+GGG G + R+ +N + ES MAFNT Y DTGLFG+Y +A +
Sbjct 369 VGGGKHV-----GSDLTQRVAINEI-----AESIMAFNTNYKDTGLFGVYAVAKADCLDD 418
Query 63 -AVKVMAEQFAGMTRVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPA 121
+ +M E RV+ ++ RA+N +KSS+ ++++ + EDIGRQLL GR I A
Sbjct 419 LSYAIMYEVTKLAYRVSDADVTRARNQLKSSLLLHMDGTSPIAEDIGRQLLTYGRRIPTA 478
Query 122 EFCRAIDNVKEADIRRVAEK-MFSKPPTVVAYGDICSVPHYEEIR 165
E ID V + ++RVA K ++ K + A G I +P Y + R
Sbjct 479 ELFARIDAVDASTVKRVANKYIYDKDIAISAIGPIQDLPDYNKFR 523
> Hs4758734
Length=489
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 48/136 (35%), Positives = 79/136 (58%), Gaps = 2/136 (1%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKS 91
S +FNT YTDTGL+GLYM+ + + + V+ +++ + T VT E+ RA+N +K+
Sbjct 347 CHSFQSFNTSYTDTGLWGLYMVCESSTVADMLHVVQKEWMRLCTSVTESEVARARNLLKT 406
Query 92 SIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEK-MFSKPPTVV 150
++ + L+ + EDIGRQ+L R I E ID V IR V K ++++ P +
Sbjct 407 NMLLQLDGSTPICEDIGRQMLCYNRRIPIPELEARIDAVNAETIREVCTKYIYNRSPAIA 466
Query 151 AYGDICSVPHYEEIRA 166
A G I +P +++IR+
Sbjct 467 AVGPIKQLPDFKQIRS 482
> 7299936
Length=382
Score = 88.2 bits (217), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 49/138 (35%), Positives = 78/138 (56%), Gaps = 2/138 (1%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANPNKASSAV-KVMAEQFAGMTRVTAEELQRAKNSIKS 91
S +FNT Y DTGL+G+Y + +P + + V E T VT E++RAKN +K+
Sbjct 240 CHSFQSFNTCYKDTGLWGIYFVCDPLQCEDMLFNVQTEWMRLCTMVTEAEVERAKNLLKT 299
Query 92 SIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEK-MFSKPPTVV 150
++ + L+ + EDIGRQ+L R I E + ID V ++R VA K ++ + P V
Sbjct 300 NMLLQLDGTTPICEDIGRQILCYNRRIPLHELEQRIDAVSVGNVRDVAMKYIYDRCPAVA 359
Query 151 AYGDICSVPHYEEIRAAL 168
A G + ++P Y IR+++
Sbjct 360 AVGPVENLPDYNRIRSSM 377
> YLR163c
Length=462
Score = 77.8 bits (190), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/139 (29%), Positives = 86/139 (61%), Gaps = 5/139 (3%)
Query 35 SAMAFNTQYTDTGLFGLYMLANPNKASSAVKV--MAEQFAGMT--RVTAEELQRAKNSIK 90
S M+F+T Y D+GL+G+Y++ + N+ + + V + +++ + +++ E+ RAK +K
Sbjct 322 SYMSFSTSYADSGLWGMYIVTDSNEHNVQLIVNEILKEWKRIKSGKISDAEVNRAKAQLK 381
Query 91 SSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAE-KMFSKPPTV 149
+++ ++L+ ++EDIGRQ++ +G+ +SP E +D + + DI A ++ +KP ++
Sbjct 382 AALLLSLDGSTAIVEDIGRQVVTTGKRLSPEEVFEQVDKITKDDIIMWANYRLQNKPVSM 441
Query 150 VAYGDICSVPHYEEIRAAL 168
VA G+ +VP+ I L
Sbjct 442 VALGNTSTVPNVSYIEEKL 460
> Hs4507843
Length=453
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 39/129 (30%), Positives = 72/129 (55%), Gaps = 2/129 (1%)
Query 38 AFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMTR--VTAEELQRAKNSIKSSIYM 95
AFN Y+D+GLFG+Y ++ A +K Q + + ++ ++Q AKN +K+ M
Sbjct 325 AFNASYSDSGLFGIYTISQATAAGDVIKAAYNQVKRIAQGNLSNTDVQAAKNKLKAGYLM 384
Query 96 NLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVVAYGDI 155
++E+ +E++G Q L++G + P+ + ID+V ADI A+K S ++ A G++
Sbjct 385 SVESSECFLEEVGSQALVAGSYMPPSTVLQQIDSVANADIINAAKKFVSGQKSMAASGNL 444
Query 156 CSVPHYEEI 164
P +E+
Sbjct 445 GHTPFVDEL 453
> Hs4507841
Length=480
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 43/138 (31%), Positives = 71/138 (51%), Gaps = 2/138 (1%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKS 91
+S F+ Y +TGL G + + + K + V+ Q+ + T T E+ R KN +++
Sbjct 338 CQSFQTFSICYAETGLLGAHFVCDRMKIDDMMFVLQGQWMRLCTSATESEVARGKNILRN 397
Query 92 SIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEK-MFSKPPTVV 150
++ +L+ V EDIGR LL GR I AE+ I V + +R + K ++ + P V
Sbjct 398 ALVSHLDGTTPVCEDIGRSLLTYGRRIPLAEWESRIAEVDASVVREICSKYIYDQCPAVA 457
Query 151 AYGDICSVPHYEEIRAAL 168
YG I +P Y IR+ +
Sbjct 458 GYGPIEQLPDYNRIRSGM 475
> CE03806
Length=485
Score = 65.1 bits (157), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 59/112 (52%), Gaps = 1/112 (0%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKS 91
+E +FNT Y +TGL G Y +A P + + + +Q+ + + + RAK S+ +
Sbjct 344 IEVFQSFNTCYKETGLVGTYFVAAPESIDNLIDSVLQQWVWLANNIDEAAVDRAKRSLHT 403
Query 92 SIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMF 143
++ + L+ V EDIGRQLL GR I E I+++ +R V ++F
Sbjct 404 NLLLMLDGSTPVCEDIGRQLLCYGRRIPTPELHARIESITVQQLRDVCRRVF 455
> CE11226
Length=471
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 39/141 (27%), Positives = 68/141 (48%), Gaps = 5/141 (3%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANP---NKASSAVKVMAEQFAGM-TRVTAEELQRAKNS 88
V + FN Y DTGLFG+Y +A+ N S +K +A ++ + + T EE+ AKN
Sbjct 326 VHNLQHFNINYKDTGLFGIYFVADAHDLNDTSGIMKSVAHEWKHLASAATEEEVAMAKNQ 385
Query 89 IKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRR-VAEKMFSKPP 147
++++Y NLE ++LL +G + +E I V +R ++ ++ +
Sbjct 386 FRTNLYQNLETNTQKAGFNAKELLYTGNLRQLSELEAQIQKVDAGAVREAISRHVYDRDL 445
Query 148 TVVAYGDICSVPHYEEIRAAL 168
V G + P+Y RA +
Sbjct 446 AAVGVGRTEAFPNYALTRAGM 466
> 7294089
Length=440
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/130 (25%), Positives = 65/130 (50%), Gaps = 1/130 (0%)
Query 35 SAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMTRVTAEELQRAKNSIKSSIY 94
S A N Y+D GLFG + A+ V+ + + V+ +++ R K +K+ I
Sbjct 311 SVKAVNASYSDAGLFGFVVSADSKDIGKTVEFLVRGLKSAS-VSDKDVARGKALLKARII 369
Query 95 MNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVVAYGD 154
+ G ++++IGRQ ++ V+ AID + ++ ++ A+K+ S V A G
Sbjct 370 SRYSSDGGLIKEIGRQAALTRNVLEADALLGAIDGISQSQVQEAAKKVGSSKLAVGAIGH 429
Query 155 ICSVPHYEEI 164
+ +VP+ ++
Sbjct 430 LANVPYASDL 439
> CE20123
Length=410
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 37/149 (24%), Positives = 69/149 (46%), Gaps = 7/149 (4%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
+ + ++ LN K A++ N Y D+GL G+ A + + K +A + +
Sbjct 269 QAVVAQILLNAAQKV--TSEAISVNVNYQDSGLVGVQFAACNTQITQVTKSIA---SAIK 323
Query 76 RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADI 135
A+ L AKN+ + + ++ V + Q+L +G +SP EF AI V D+
Sbjct 324 SAKADGLDNAKNTAAVQVLSDAQHASEVALEKATQVL-AGVEVSPREFADAIRAVTAQDV 382
Query 136 RRVAEKMFSKPPTVVAYGDICSVPHYEEI 164
+ ++ K ++ AYG VP+ +E+
Sbjct 383 TQALSRVNGK-LSLAAYGSTSLVPYLDEL 410
> CE19592
Length=427
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 61/128 (47%), Gaps = 8/128 (6%)
Query 38 AFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMTRVT-AEELQRAKNSIKSSIYMN 96
AF Y +GL G+Y+LA A SAV+ + + R T ++++ K + I N
Sbjct 307 AFQAPYDGSGLVGVYLLATGANADSAVRATVK----VLRTTQVQDIEGCKRRAIADILFN 362
Query 97 LENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVVAYGDIC 156
EN D+ L +G +E I ++E+D+++ + F + + AYG+
Sbjct 363 AENNVYSAYDLATNALYNGP--EQSELIAEIQKIQESDVQKYTKAAFER-LAISAYGNYG 419
Query 157 SVPHYEEI 164
+P+ +E+
Sbjct 420 RIPYADEL 427
> SPCC613.10
Length=426
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 34/147 (23%), Positives = 65/147 (44%), Gaps = 27/147 (18%)
Query 31 DWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMTRVTAEELQRAKNSIK 90
++ +A+A T Y+D L + + + KA + TA E +A S+
Sbjct 294 EYKATAVADLTPYSDASLLSVVISGSCPKA--------------IKATASESFKALKSLS 339
Query 91 SSIYMNLENRGIVM---------EDIGRQLLMSGRVISPAE----FCRAIDNVKEADIRR 137
S+I ++ GI M E + + + ++S ++ F D V A I +
Sbjct 340 SNIPNDVVKSGIAMAKTKYLSAFEPVTLNAISASSLVSASKGSDAFISGFDKVTPASISK 399
Query 138 VAEKMFSKPPTVVAYGDICSVPHYEEI 164
V + +KP + VA G++ +P+Y+E+
Sbjct 400 VVSSLLAKPASTVAVGNLDVLPYYDEL 426
> CE23962
Length=422
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 34/132 (25%), Positives = 68/132 (51%), Gaps = 13/132 (9%)
Query 35 SAMAFNTQYTDTGLFGLYMLANPNKASSAVK--VMAEQFAGMTRVTAEELQRAKNSIKSS 92
SA AF + D+GL G+Y++ ++A+ AV V A + + + A + Q N++++S
Sbjct 302 SASAFQAVHADSGLAGVYLVVEGSQANQAVSNVVGALKSLKVADIEAVKKQAFNNALRAS 361
Query 93 IYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVVAY 152
+ +N I + QL S + + I NV +D+ A+K+ +K ++ +Y
Sbjct 362 AHS--DNFAI---ERASQLFQ-----SQDNYIQQIPNVSASDVEVAAKKLTTK-LSLASY 410
Query 153 GDICSVPHYEEI 164
G++ VP+ + +
Sbjct 411 GNVSEVPYVDTL 422
> YBL045c
Length=457
Score = 37.7 bits (86), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 36/165 (21%), Positives = 76/165 (46%), Gaps = 5/165 (3%)
Query 7 GAFSTGGPGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLA-NPNKASSAVK 65
G+++ P + L+ + +Y ++ F+ Y D+GL+G N +
Sbjct 286 GSYNAFEPASRLQGIKLLDNIQEYQLCDNFNHFSLSYKDSGLWGFSTATRNVTMIDDLIH 345
Query 66 VMAEQFAGMT-RVTAEELQRAKNSIKSSIYMNLENRGIVMED--IGRQLLMSGRVISPAE 122
+Q+ +T VT E++RAK+ +K + E+ V + +G ++L+ G +S E
Sbjct 346 FTLKQWNRLTISVTDTEVERAKSLLKLQLGQLYESGNPVNDANLLGAEVLIKGSKLSLGE 405
Query 123 FCRAIDNVKEADIRRVA-EKMFSKPPTVVAYGDICSVPHYEEIRA 166
+ ID + D++ A ++++ + + G I + Y IR+
Sbjct 406 AFKKIDAITVKDVKAWAGKRLWDQDIAIAGTGQIEGLLDYMRIRS 450
> Hs5174623
Length=369
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 34/70 (48%), Gaps = 8/70 (11%)
Query 77 VTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIR 136
+T EE+Q SI IY N+ EDI +L S ISP+E +D +
Sbjct 95 ITKEEIQ----SISEKIYRADTNKA-QKEDI---VLNSQNCISPSETRNQVDRCPKPLFT 146
Query 137 RVAEKMFSKP 146
V EK+FSKP
Sbjct 147 YVNEKLFSKP 156
> At5g18510
Length=702
Score = 33.1 bits (74), Expect = 0.31, Method: Composition-based stats.
Identities = 26/106 (24%), Positives = 45/106 (42%), Gaps = 1/106 (0%)
Query 48 LFGLYMLANPNKASSAVKVMAEQFAGMTRVTAEELQRAKNSIKSSIYMNLENRGIVMEDI 107
L G +L +P + M + + +V + L +AK + S + RG ME
Sbjct 129 LLGFSVLGSPVFSPLECSEMRDSAEKLEKVRRDSLGKAKRVSQRSWTSSFMGRGGQMEHE 188
Query 108 GRQLLMSGRVISPAEFCRAID-NVKEADIRRVAEKMFSKPPTVVAY 152
+L + P +FCR+I NV +R + + P V+A+
Sbjct 189 AFLVLWLSLFVFPGKFCRSISTNVIPIAVRLARGERIALAPAVLAF 234
> At2g41220
Length=1629
Score = 31.6 bits (70), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 35/68 (51%), Gaps = 2/68 (2%)
Query 4 GGGGAFSTGGPGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSA 63
GG G+ +T G G G+ T + ++ N++ + +F+ +T G+ L++ + N A
Sbjct 139 GGCGSDNTSGDGSGLMTSIPWDLFNEWAEKQGIASFDRTHTGVGM--LFLPRDDNIRKEA 196
Query 64 VKVMAEQF 71
KV+ F
Sbjct 197 KKVITSIF 204
> CE24908
Length=511
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 128 DNVKEADIRRVAEKMFSKPPTVVAYGDICSV 158
+N++ A I +KM+ PP V A+ +IC +
Sbjct 408 NNIRNATIDDTIQKMYYSPPVVKAFENICFI 438
> At5g23110
Length=4706
Score = 28.5 bits (62), Expect = 6.7, Method: Composition-based stats.
Identities = 28/80 (35%), Positives = 41/80 (51%), Gaps = 6/80 (7%)
Query 58 NKASSAVKVMAEQFAGMTRVTAEELQRAKNSIKSS--IYMNLENRGIVMEDIGRQ-LLMS 114
NK SS+ V Q RVTA+EL A + + S+ I M LEN+ ++ + Q L
Sbjct 4576 NKTSSSQPVNEMQLG---RVTAKELVEAVHEMLSAAGINMELENQSLLQRTLTLQEELKD 4632
Query 115 GRVISPAEFCRAIDNVKEAD 134
+V E RA ++KEA+
Sbjct 4633 SKVAFLLEQERAEASMKEAE 4652
Lambda K H
0.318 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2671071884
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40