bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_1668_orf2
Length=184
Score E
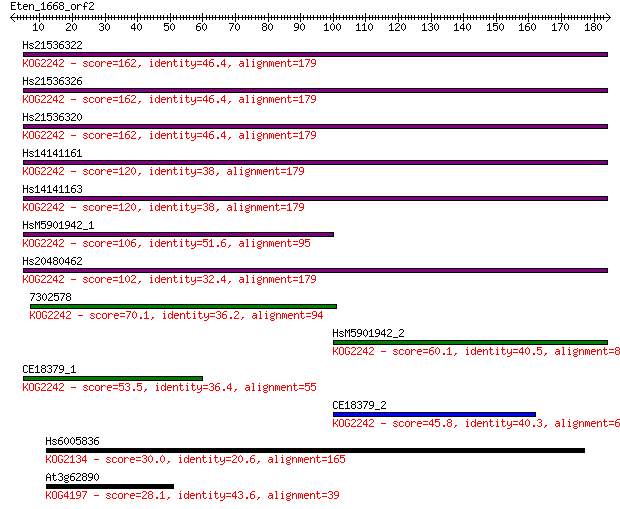
Sequences producing significant alignments: (Bits) Value
Hs21536322 162 3e-40
Hs21536326 162 4e-40
Hs21536320 162 5e-40
Hs14141161 120 2e-27
Hs14141163 120 2e-27
HsM5901942_1 106 3e-23
Hs20480462 102 3e-22
7302578 70.1 3e-12
HsM5901942_2 60.1 2e-09
CE18379_1 53.5 2e-07
CE18379_2 45.8 5e-05
Hs6005836 30.0 3.1
At3g62890 28.1 9.9
> Hs21536322
Length=804
Score = 162 bits (410), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 83/179 (46%), Positives = 114/179 (63%), Gaps = 3/179 (1%)
Query 5 PRSVSECEFIMMCGLPACGKTYWIERHIEKYPKKSYVLLGTNAVIDQMKVMGLKRQANYA 64
P+S +ECE +MM GLPA GKT W +H P K Y +LGTNA++D+M+VMGL+RQ NYA
Sbjct 415 PKSKAECEILMMVGLPAAGKTTWAIKHAASNPSKKYNILGTNAIMDKMRVMGLRRQRNYA 474
Query 65 NRWEELMSTATEVFNSLVAYAGSGVVPRNVIIDQTNVFKRARARKVQPFLGWGTRRCVVM 124
RW+ L+ AT+ N L+ A RN I+DQTNV+ A+ RK++PF G+ + V+
Sbjct 475 GRWDVLIQQATQCLNRLIQIAARKK--RNYILDQTNVYGSAQRRKMRPFEGFQRKAIVIC 532
Query 125 VTDEPTLQQRTQKREREEGKMVPVAAVMDMKAAFALPSTDGGFTVIDYVEMPEGESSKV 183
TDE L+ RT KR EEGK VP AV++MKA F LP + ++E+ E+ K+
Sbjct 533 PTDE-DLKDRTIKRTDEEGKDVPDHAVLEMKANFTLPDVGDFLDEVLFIELQREEADKL 590
> Hs21536326
Length=856
Score = 162 bits (410), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 83/179 (46%), Positives = 114/179 (63%), Gaps = 3/179 (1%)
Query 5 PRSVSECEFIMMCGLPACGKTYWIERHIEKYPKKSYVLLGTNAVIDQMKVMGLKRQANYA 64
P+S +ECE +MM GLPA GKT W +H P K Y +LGTNA++D+M+VMGL+RQ NYA
Sbjct 415 PKSKAECEILMMVGLPAAGKTTWAIKHAASNPSKKYNILGTNAIMDKMRVMGLRRQRNYA 474
Query 65 NRWEELMSTATEVFNSLVAYAGSGVVPRNVIIDQTNVFKRARARKVQPFLGWGTRRCVVM 124
RW+ L+ AT+ N L+ A RN I+DQTNV+ A+ RK++PF G+ + V+
Sbjct 475 GRWDVLIQQATQCLNRLIQIAAR--KKRNYILDQTNVYGSAQRRKMRPFEGFQRKAIVIC 532
Query 125 VTDEPTLQQRTQKREREEGKMVPVAAVMDMKAAFALPSTDGGFTVIDYVEMPEGESSKV 183
TDE L+ RT KR EEGK VP AV++MKA F LP + ++E+ E+ K+
Sbjct 533 PTDE-DLKDRTIKRTDEEGKDVPDHAVLEMKANFTLPDVGDFLDEVLFIELQREEADKL 590
> Hs21536320
Length=756
Score = 162 bits (409), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 83/179 (46%), Positives = 114/179 (63%), Gaps = 3/179 (1%)
Query 5 PRSVSECEFIMMCGLPACGKTYWIERHIEKYPKKSYVLLGTNAVIDQMKVMGLKRQANYA 64
P+S +ECE +MM GLPA GKT W +H P K Y +LGTNA++D+M+VMGL+RQ NYA
Sbjct 315 PKSKAECEILMMVGLPAAGKTTWAIKHAASNPSKKYNILGTNAIMDKMRVMGLRRQRNYA 374
Query 65 NRWEELMSTATEVFNSLVAYAGSGVVPRNVIIDQTNVFKRARARKVQPFLGWGTRRCVVM 124
RW+ L+ AT+ N L+ A RN I+DQTNV+ A+ RK++PF G+ + V+
Sbjct 375 GRWDVLIQQATQCLNRLIQIAAR--KKRNYILDQTNVYGSAQRRKMRPFEGFQRKAIVIC 432
Query 125 VTDEPTLQQRTQKREREEGKMVPVAAVMDMKAAFALPSTDGGFTVIDYVEMPEGESSKV 183
TDE L+ RT KR EEGK VP AV++MKA F LP + ++E+ E+ K+
Sbjct 433 PTDE-DLKDRTIKRTDEEGKDVPDHAVLEMKANFTLPDVGDFLDEVLFIELQREEADKL 490
> Hs14141161
Length=806
Score = 120 bits (300), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 68/179 (37%), Positives = 97/179 (54%), Gaps = 4/179 (2%)
Query 5 PRSVSECEFIMMCGLPACGKTYWIERHIEKYPKKSYVLLGTNAVIDQMKVMGLKRQANYA 64
P +CE +MM GLP GKT W+ +H + P K Y +LGTN ++D+M V G K+Q
Sbjct 472 PEEKKDCEVVMMIGLPGAGKTTWVTKHAAENPGK-YNILGTNTIMDKMMVAGFKKQMADT 530
Query 65 NRWEELMSTATEVFNSLVAYAGSGVVPRNVIIDQTNVFKRARARKVQPFLGWGTRRCVVM 124
+ L+ A + + A RN I+DQTNV A+ RK+ F G+ R+ VV+
Sbjct 531 GKLNTLLQRAPQCLGKFIEIAAR--KKRNFILDQTNVSAAAQRRKMCLFAGF-QRKAVVV 587
Query 125 VTDEPTLQQRTQKREREEGKMVPVAAVMDMKAAFALPSTDGGFTVIDYVEMPEGESSKV 183
+ +QRTQK+ EGK +P AV+ MK F LP F I YVE+ + E+ K+
Sbjct 588 CPKDEDYKQRTQKKAEVEGKDLPEHAVLKMKGNFTLPEVAECFDEITYVELQKEEAQKL 646
> Hs14141163
Length=824
Score = 120 bits (300), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 68/179 (37%), Positives = 97/179 (54%), Gaps = 4/179 (2%)
Query 5 PRSVSECEFIMMCGLPACGKTYWIERHIEKYPKKSYVLLGTNAVIDQMKVMGLKRQANYA 64
P +CE +MM GLP GKT W+ +H + P K Y +LGTN ++D+M V G K+Q
Sbjct 490 PEEKKDCEVVMMIGLPGAGKTTWVTKHAAENPGK-YNILGTNTIMDKMMVAGFKKQMADT 548
Query 65 NRWEELMSTATEVFNSLVAYAGSGVVPRNVIIDQTNVFKRARARKVQPFLGWGTRRCVVM 124
+ L+ A + + A RN I+DQTNV A+ RK+ F G+ R+ VV+
Sbjct 549 GKLNTLLQRAPQCLGKFIEIAAR--KKRNFILDQTNVSAAAQRRKMCLFAGF-QRKAVVV 605
Query 125 VTDEPTLQQRTQKREREEGKMVPVAAVMDMKAAFALPSTDGGFTVIDYVEMPEGESSKV 183
+ +QRTQK+ EGK +P AV+ MK F LP F I YVE+ + E+ K+
Sbjct 606 CPKDEDYKQRTQKKAEVEGKDLPEHAVLKMKGNFTLPEVAECFDEITYVELQKEEAQKL 664
> HsM5901942_1
Length=507
Score = 106 bits (264), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 49/95 (51%), Positives = 64/95 (67%), Gaps = 2/95 (2%)
Query 5 PRSVSECEFIMMCGLPACGKTYWIERHIEKYPKKSYVLLGTNAVIDQMKVMGLKRQANYA 64
P+S +ECE +MM GLPA GKT W +H P K Y +LGTNA++D+M+VMGL+RQ NYA
Sbjct 415 PKSKAECEILMMVGLPAAGKTTWAIKHAASNPSKKYNILGTNAIMDKMRVMGLRRQRNYA 474
Query 65 NRWEELMSTATEVFNSLVAYAGSGVVPRNVIIDQT 99
RW+ L+ AT+ N L+ A RN I+DQT
Sbjct 475 GRWDVLIQQATQCLNRLIQIAAR--KKRNYILDQT 507
> Hs20480462
Length=444
Score = 102 bits (255), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 58/179 (32%), Positives = 101/179 (56%), Gaps = 3/179 (1%)
Query 5 PRSVSECEFIMMCGLPACGKTYWIERHIEKYPKKSYVLLGTNAVIDQMKVMGLKRQANYA 64
P+++ ECE I+M GLP GKT W ++ ++ P+K Y +LG V++QM++ GL+
Sbjct 143 PKTIEECEVILMVGLPGSGKTQWALKYAKENPEKRYNVLGAETVLNQMRMKGLEEPEMDP 202
Query 65 NRWEELMSTATEVFNSLVAYAGSGVVPRNVIIDQTNVFKRARARKVQPFLGWGTRRCVVM 124
+ L+ A++ + LV A RN I+DQ NV+ + RK+ F + +R+ VV+
Sbjct 203 KSRDLLVQQASQCLSKLVQIASR--TKRNFILDQCNVYNSGQRRKLLLFKTF-SRKVVVV 259
Query 125 VTDEPTLQQRTQKREREEGKMVPVAAVMDMKAAFALPSTDGGFTVIDYVEMPEGESSKV 183
V +E ++R + R+ EG VP + +++MKA F+LP + Y E+ + E+ +
Sbjct 260 VPNEEDWKKRLELRKEVEGDDVPESIMLEMKANFSLPEKCDYMDEVTYGELEKEEAQPI 318
> 7302578
Length=795
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 34/95 (35%), Positives = 51/95 (53%), Gaps = 4/95 (4%)
Query 7 SVSECEFIMMCGLPACGKTYWIERHIEKYPKKSYVLLGTNAVIDQMKVMGLKRQANYANR 66
S ECE I++ GLP GKT+W +H+ + K Y L+G +A I +M + G R+ + R
Sbjct 703 SRKECEVILLVGLPGAGKTHWAHKHVAENADKRYELIGPDAFISKMTIDGASRKTVHKGR 762
Query 67 WEELMSTATEVFNSLVAYAGSGVV-PRNVIIDQTN 100
W+++ NSL A + RN I+DQ N
Sbjct 763 WDKVYEI---CLNSLAALEDIAMKRRRNFILDQVN 794
> HsM5901942_2
Length=349
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/84 (40%), Positives = 50/84 (59%), Gaps = 1/84 (1%)
Query 100 NVFKRARARKVQPFLGWGTRRCVVMVTDEPTLQQRTQKREREEGKMVPVAAVMDMKAAFA 159
NV+ A+ RK++PF G+ + V+ TDE L+ RT KR EEGK VP AV++MKA F
Sbjct 1 NVYGSAQRRKMRPFEGFQRKAIVICPTDE-DLKDRTIKRTDEEGKDVPDHAVLEMKANFT 59
Query 160 LPSTDGGFTVIDYVEMPEGESSKV 183
LP + ++E+ E+ K+
Sbjct 60 LPDVGDFLDEVLFIELQREEADKL 83
> CE18379_1
Length=503
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 34/55 (61%), Gaps = 0/55 (0%)
Query 5 PRSVSECEFIMMCGLPACGKTYWIERHIEKYPKKSYVLLGTNAVIDQMKVMGLKR 59
P + +C +M GLP GKT W+ R++ ++P + + L+ + V+D MKV G+ R
Sbjct 410 PAAKKDCTVLMTVGLPGVGKTTWVRRYLAEHPHEHWTLISADVVMDAMKVNGVPR 464
> CE18379_2
Length=414
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 36/62 (58%), Gaps = 1/62 (1%)
Query 100 NVFKRARARKVQPFLGWGTRRCVVMVTDEPTLQQRTQKREREEGKMVPVAAVMDMKAAFA 159
N F R RK+ F + R+C++ V DE T Q+R K+E EE + + A+M KAA +
Sbjct 1 NCFAEKRKRKLMAFEEFN-RKCMLFVPDEDTHQKRLIKQEHEENEKLDTDAMMQHKAAMS 59
Query 160 LP 161
LP
Sbjct 60 LP 61
> Hs6005836
Length=521
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 34/168 (20%), Positives = 63/168 (37%), Gaps = 34/168 (20%)
Query 12 EFIMMCGLPACGKTYWIERHIEKYPKKSYVLLGTNAVIDQMKVMGLKRQANYANRWEELM 71
E ++ G P GK+ ++++H+ YV + + + W+ +
Sbjct 366 EVVVAVGFPGAGKSTFLKKHLVS---AGYVHVNRDTL----------------GSWQRCV 406
Query 72 STATEVFNSLVAYAGSGVVPRNVIIDQTNVFKRARARKVQPFLGWGTR-RCVVMVTDEPT 130
+T + V ID TN +RAR VQ G RC +
Sbjct 407 TTCETALKQ----------GKRVAIDNTNPDAASRARYVQCARAAGVPCRCFLFTATLEQ 456
Query 131 LQQRTQKREREEGKMVPVAAVM--DMKAAFALPSTDGGFTVIDYVEMP 176
+ + RE + +PV+ ++ + F P+ GF+ I +E+P
Sbjct 457 ARHNNRFREMTDSSHIPVSDMVMYGYRKQFEAPTLAEGFSAI--LEIP 502
> At3g62890
Length=558
Score = 28.1 bits (61), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 25/45 (55%), Gaps = 7/45 (15%)
Query 12 EFIMMCGLPACGKT------YWIERHIEKYPKKSYVLLGTNAVID 50
EF M L ACG+ W+ +I+KY + ++LGT A+ID
Sbjct 199 EFTMSTVLSACGRLGALEQGKWVHAYIDKYHVEIDIVLGT-ALID 242
Lambda K H
0.319 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2950576972
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40