bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
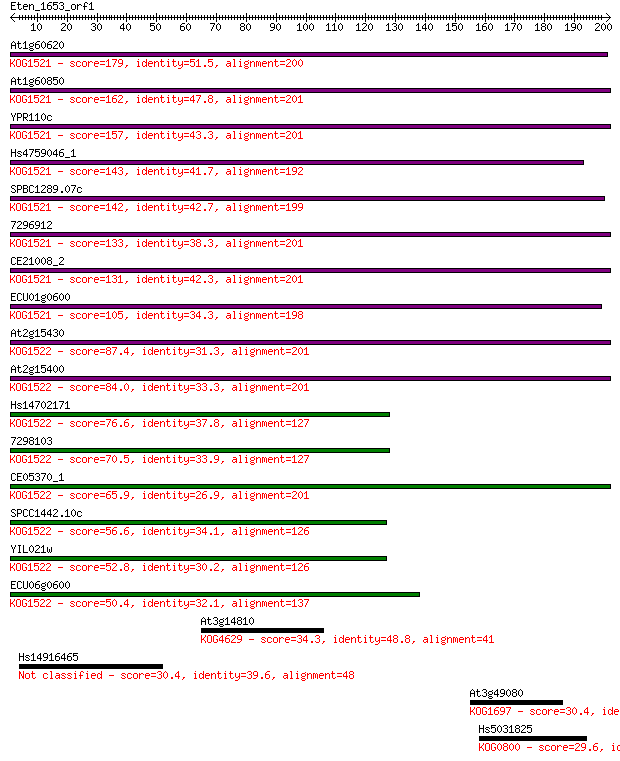
Query= Eten_1653_orf1
Length=201
Score E
Sequences producing significant alignments: (Bits) Value
At1g60620 179 3e-45
At1g60850 162 4e-40
YPR110c 157 1e-38
Hs4759046_1 143 3e-34
SPBC1289.07c 142 3e-34
7296912 133 3e-31
CE21008_2 131 1e-30
ECU01g0600 105 7e-23
At2g15430 87.4 2e-17
At2g15400 84.0 2e-16
Hs14702171 76.6 3e-14
7298103 70.5 2e-12
CE05370_1 65.9 6e-11
SPCC1442.10c 56.6 4e-08
YIL021w 52.8 4e-07
ECU06g0600 50.4 2e-06
At3g14810 34.3 0.15
Hs14916465 30.4 2.3
At3g49080 30.4 2.8
Hs5031825 29.6 4.4
> At1g60620
Length=385
Score = 179 bits (454), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 103/229 (44%), Positives = 137/229 (59%), Gaps = 34/229 (14%)
Query 1 EVLAHRLGLIPFKVNPKKLKFRNPNQELSEENSLKFKMHVKCSPGDLKPHQKSMPVYARD 60
EVLAHRLGLIP +P+ ++ + N + +E+N++ FK+HVKC GD P +K V +
Sbjct 136 EVLAHRLGLIPIAADPRLFEYLSENDQPNEKNTIVFKLHVKCLKGD--PRRK---VLTSE 190
Query 61 LKWIPM-SERQKQ----------------------KFAQDPPAPVHPDILITKLRPGQEI 97
LKW+P SE K+ +FA++P P DILI KL PGQEI
Sbjct 191 LKWLPNGSELIKESGGSTTTPKTYTSFNHSQDSFPEFAENPIRPTLKDILIAKLGPGQEI 250
Query 98 ELFGFLEKGLGKTHAKWSPVATAVYRLEPEFVFTSPIEGEEAKELKELCPMGVFDIED-- 155
EL KG+GKTHAKWSPVATA YR+ PE V EG+ A+EL ++CP VFDIED
Sbjct 251 ELEAHAVKGIGKTHAKWSPVATAWYRMLPEVVLLKEFEGKHAEELVKVCPKKVFDIEDMG 310
Query 156 -STGRAYAKFPRNCTTCRACLE---RFENQLQLNKIPDQFIFSIESTGS 200
RA PR+C+ CR C+ +E+Q+ L ++ + FIF+IESTGS
Sbjct 311 QGRKRATVARPRDCSLCRECIRDGVEWEDQVDLRRVKNHFIFTIESTGS 359
> At1g60850
Length=375
Score = 162 bits (410), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 96/229 (41%), Positives = 126/229 (55%), Gaps = 33/229 (14%)
Query 1 EVLAHRLGLIPFKVNPKKLKFRNPNQELSEENSLKFKMHVKCSPGDLKPHQKSMPVYARD 60
EVLAHR+GLIP +P+ ++ + + + +E+N++ FK+HVKC +P K V D
Sbjct 129 EVLAHRMGLIPIAADPRLFEYLSEHDQANEKNTIVFKLHVKCPKN--RPRLK---VLTSD 183
Query 61 LKWIPM-------SERQKQK----------------FAQDPPAPVHPDILITKLRPGQEI 97
LKW+P SE + K FA +P P DILI KL PGQEI
Sbjct 184 LKWLPNGSELLRESENKTSKPKTYTSFSCSQDSLPEFANNPITPCDLDILIAKLAPGQEI 243
Query 98 ELFGFLEKGLGKTHAKWSPVATAVYRLEPEFVFTSPIEGEEAKELKELCPMGVFDIED-- 155
EL KG+GKTHAKWSPV TA YR+ PE V +E E A+ L +CP VFDIED
Sbjct 244 ELEAHAVKGIGKTHAKWSPVGTAWYRMHPEVVLRGEVEDELAERLVNVCPQNVFDIEDMG 303
Query 156 -STGRAYAKFPRNCTTCRACLE--RFENQLQLNKIPDQFIFSIESTGSV 201
RA PR CT C+ C+ + + L + + FIF+IESTGS+
Sbjct 304 KGKKRATVAQPRKCTLCKECVRDDDLVDHVDLGSVKNHFIFNIESTGSL 352
> YPR110c
Length=335
Score = 157 bits (397), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 87/215 (40%), Positives = 126/215 (58%), Gaps = 15/215 (6%)
Query 1 EVLAHRLGLIPFKVNPKKLKFRNPN----QELSEENSLKFKMHVKCSPGDLKPHQKSMP- 55
EVLAHR+GL+P KV+P L + + N ++ ++EN++ ++VKC+ P + P
Sbjct 95 EVLAHRIGLVPLKVDPDMLTWVDSNLPDDEKFTDENTIVLSLNVKCTRNPDAPKGSTDPK 154
Query 56 -------VYARDLKWIPMSERQKQKFAQDPPAPVHPDILITKLRPGQEIELFGFLEKGLG 108
VYARDLK+ P RQ FA P P PDIL+ KLRPGQEI L G+G
Sbjct 155 ELYNNAHVYARDLKFEPQG-RQSTTFADCPVVPADPDILLAKLRPGQEISLKAHCILGIG 213
Query 109 KTHAKWSPVATAVYRLEPEFVFTSPIEGEEAKELKELCPMGVFDIEDSTGRAYAKFPRNC 168
HAK+SPV+TA YRL P+ PI+GE A+ ++ P GV I++ + AY K R
Sbjct 214 GDHAKFSPVSTASYRLLPQINILQPIKGESARRFQKCFPPGVIGIDEGSDEAYVKDARKD 273
Query 169 TTCRACL--ERFENQLQLNKIPDQFIFSIESTGSV 201
T R L E F ++++L ++ + FIF++ES G++
Sbjct 274 TVSREVLRYEEFADKVKLGRVRNHFIFNVESAGAM 308
> Hs4759046_1
Length=314
Score = 143 bits (360), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 80/205 (39%), Positives = 118/205 (57%), Gaps = 14/205 (6%)
Query 1 EVLAHRLGLIPFKVNPKKLKFRNP-NQELSEENSLKFKMHVKCSPGDLKPHQKSMP---- 55
E+LAHRLGLIP +P+ ++RN ++E +E ++L+F++ V+C+ S P
Sbjct 104 EILAHRLGLIPIHADPRLFEYRNQGDEEGTEIDTLQFRLQVRCTRNPHAAKDSSDPNELY 163
Query 56 ----VYARDLKWIPMSERQKQKFAQDPPAPVHPDILITKLRPGQEIELFGFLEKGLGKTH 111
VY R + WIP+ Q F + PVH DILI +LRPGQEI+L KG+GK H
Sbjct 164 VNHKVYTRHMTWIPLGN-QADLFPEGTIRPVHDDILIAQLRPGQEIDLLMHCVKGIGKDH 222
Query 112 AKWSPVATAVYRLEPEFVFTSPIEGEEAKELKELCPMGVFDIEDSTGRAYAKF--PRNCT 169
AK+SPVATA YRL P+ P+EGE A+EL GV ++++ G+ A+ PR T
Sbjct 223 AKFSPVATASYRLLPDITLLEPVEGEAAEELSRCFSPGVIEVQEVQGKKVARVANPRLDT 282
Query 170 TCRACL--ERFENQLQLNKIPDQFI 192
R E+ + ++L ++ D +I
Sbjct 283 FSREIFRNEKLKKVVRLARVRDHYI 307
> SPBC1289.07c
Length=348
Score = 142 bits (359), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 85/217 (39%), Positives = 125/217 (57%), Gaps = 19/217 (8%)
Query 1 EVLAHRLGLIPFKVNPKKLKFRN---PNQELS--EENSLKFKMHVKC--------SPGDL 47
EVL+HR+GL+P +P K+ P QE + + +++ F ++ KC D
Sbjct 101 EVLSHRIGLVPISADPDMFKWFQHPLPGQEATHTDYDTVVFSLNKKCEFNKNAATDEKDP 160
Query 48 KPHQKSMPVYARDLKWIPMSERQKQKFAQDPPAPVHPDILITKLRPGQEIELFGFLEKGL 107
K + VY+ DL W P RQ+++FA +P V+PDI++ KLRPGQEI+L G+
Sbjct 161 KRLYVNSEVYSGDLIWKPQG-RQEERFADNPIRVVNPDIVVAKLRPGQEIDLEAHAILGI 219
Query 108 GKTHAKWSPVATAVYRLEPEFVFTSPIEGEEAKELKELCPMGVFDIE---DSTGRAYAKF 164
G+ HAK+SPVATA YRL P SPIEGE+A + ++ P GV ++E D +A
Sbjct 220 GQDHAKFSPVATASYRLLPTIHILSPIEGEDAVKFQKCFPKGVIELEEGPDGKKQARVAD 279
Query 165 PRNCTTCRACLER--FENQLQLNKIPDQFIFSIESTG 199
R T R CL F +++QL ++ D ++FS+ESTG
Sbjct 280 VRKDTVSRECLRHPEFADKVQLGRVRDHYLFSVESTG 316
> 7296912
Length=333
Score = 133 bits (334), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 77/213 (36%), Positives = 117/213 (54%), Gaps = 14/213 (6%)
Query 1 EVLAHRLGLIPFKVNPKKLKFRN--PNQELSEENSLKFKMHVKCS--------PGDLKPH 50
EVLAHR+GL+P + +P+ +R + +E+++L+F++ VKCS +
Sbjct 96 EVLAHRMGLLPLRADPRLFAYRTEESTEAGTEQDTLEFELKVKCSRRRDAGKDQSNFDDI 155
Query 51 QKSMPVYARDLKWIPMSERQKQKFAQDPPAPVHPDILITKLRPGQEIELFGFLEKGLGKT 110
K+ VY+ LKW+P +Q Q +++ +H DILI +LRPG E++L KGLG+
Sbjct 156 YKNHKVYSGHLKWLPKG-KQAQIYSESAVNCIHDDILIAQLRPGHELDLRLVAVKGLGRD 214
Query 111 HAKWSPVATAVYRLEPEFVFTSPIEGEEAKELKELCPMGVFDIEDSTGRAYAKFPRNCTT 170
HAK+SPVATA YRL P + G++A L+ GV I D AY K R T
Sbjct 215 HAKFSPVATATYRLLPMIKLNREVTGKDAYLLQNCFSPGVIGI-DENETAYVKDARYDTC 273
Query 171 CRACLE--RFENQLQLNKIPDQFIFSIESTGSV 201
R + + + L +I D +IFS+ES G++
Sbjct 274 SRNVYRYPQLNDAVTLARIRDHYIFSVESVGAL 306
> CE21008_2
Length=362
Score = 131 bits (329), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 85/227 (37%), Positives = 117/227 (51%), Gaps = 26/227 (11%)
Query 1 EVLAHRLGLIPFKVNPKKLKF--------RNPNQELSEE------NSLKFKMHVKCS--- 43
EVL HRLGL+P +V+P+ +F + EE +L FK++V C+
Sbjct 107 EVLCHRLGLLPLRVDPRGFQFPKEKVVGINEKGVDCDEEPEGDPAKNLIFKINVSCTKNR 166
Query 44 ---PGDLKPHQ--KSMPVYARDLKWIPMSERQKQKFAQDPPAPVHPDILITKLRPGQEIE 98
P P Q + VY+R +W+P+++++ Q + P V DIL+ KLRPGQEIE
Sbjct 167 NAVPTATDPKQLYHNSSVYSRAFEWVPIADQKTQFTEEAHPRMVSDDILVAKLRPGQEIE 226
Query 99 LFGFLEKGLGKTHAKWSPVATAVYRLEPEFVFTSPIEGEEAKELKELCPMGVFDIEDSTG 158
KG+G+ HAK+SPVATA YRL P + I GE A+ LK + GV IE
Sbjct 227 ASCHAVKGIGRDHAKFSPVATASYRLLPTIRLNAEISGEAAERLKSVFSEGVIAIEKKGA 286
Query 159 R--AYAKFPRNCTTCRACL--ERFENQLQLNKIPDQFIFSIESTGSV 201
+ A K R T R E +QL K FIFS+ESTG++
Sbjct 287 KRIAVVKDARKDTCSRNVFRHEDLSKVVQLGKNKQHFIFSVESTGAL 333
> ECU01g0600
Length=305
Score = 105 bits (261), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 68/202 (33%), Positives = 107/202 (52%), Gaps = 17/202 (8%)
Query 1 EVLAHRLGLIPFKVNPKKLKFRNPNQELSEENSLKFKMHVKCSPGDLKPHQKSMPVYARD 60
E +AHRLGL+P +V+P+ +F +EL E NSL+F + V+ + + VY+ D
Sbjct 85 EFIAHRLGLVPLEVDPEMFEF--AGEELDERNSLRFTLDVR------NESEGVLNVYSDD 136
Query 61 LKWIPMSERQKQKFAQDPPAPVHPDILITKLRPGQEIELFGFLEKGLGKTHAKWSPVATA 120
++W P+ Q++ F +L+ +L GQ+I K +GK HAKWSPV A
Sbjct 137 IRWNPVGS-QREIFRD---VKFSSRVLMFRLARGQKISAELVCTKNVGKVHAKWSPVCPA 192
Query 121 VYRLEPEFVFTSPIEGEEAKELKELCPMGVFDIEDSTGR--AYAKFPRNCTTCRACL--E 176
YRL P + I E+A +L++ GV D+ S GR AY PR + R L E
Sbjct 193 TYRLMP-LIEIGDIFDEDAYKLQKCFSEGVIDVRPSDGRMKAYVSNPRLDSMSREALRHE 251
Query 177 RFENQLQLNKIPDQFIFSIEST 198
F++++ + + D F+F+I +
Sbjct 252 EFKDKIFIGRRSDYFLFTINTV 273
> At2g15430
Length=319
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 63/216 (29%), Positives = 98/216 (45%), Gaps = 19/216 (8%)
Query 1 EVLAHRLGLIPFKV-NPKKLKFRNPNQELSEEN-----SLKFKMHVKCSPGDLKPHQKSM 54
E +AHRLGLIP ++F + S++F++ KC D S
Sbjct 65 EFIAHRLGLIPLTSERAMSMRFSRDCDACDGDGQCEFCSVEFRLSSKCVT-DQTLDVTSR 123
Query 55 PVYARDLKWIPMSERQKQKFAQDPPAPVHPDILITKLRPGQEIELFGFLEKGLGKTHAKW 114
+Y+ D P+ + + H I+I KLR GQE++L KG+GK HAKW
Sbjct 124 DLYSADPTVTPVDFTIDSSVSD---SSEHKGIIIVKLRRGQELKLRAIARKGIGKDHAKW 180
Query 115 SPVATAVYRLEPEFVFT----SPIEGEEAKELKELCPMGVFDIEDSTGRAYAKFPRNCTT 170
SP AT + EP+ + + EE +L E P VF ++ T + P T
Sbjct 181 SPAATVTFMYEPDIIINEDMMDTLSDEEKIDLIESSPTKVFGMDPVTRQVVVVDPEAYTY 240
Query 171 CRACLERFENQ-----LQLNKIPDQFIFSIESTGSV 201
+++ E ++++ D FIF++ESTG+V
Sbjct 241 DEEVIKKAEAMGKPGLIEISPKDDSFIFTVESTGAV 276
> At2g15400
Length=319
Score = 84.0 bits (206), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 67/231 (29%), Positives = 103/231 (44%), Gaps = 49/231 (21%)
Query 1 EVLAHRLGLIPFKVNPKKLKFR-------NPNQELSEENSLKFKMHVKCSPGDLKPHQKS 53
E +A RL LIP + + + R E E S++F + KC ++
Sbjct 65 EFIAQRLSLIPL-TSERAMSMRFCQDCEDCNGDEHCEFCSVEFPLSAKCVT------DQT 117
Query 54 MPVYARDLKWIPMSERQKQKFAQDPPA-PV-------------HPDILITKLRPGQEIEL 99
+ V +RDL ++ DP PV H I+I KLR GQE++L
Sbjct 118 LDVTSRDL------------YSADPTVTPVDFTSNSSTSDSSEHKGIIIAKLRRGQELKL 165
Query 100 FGFLEKGLGKTHAKWSPVATAVYRLEPEFVFT----SPIEGEEAKELKELCPMGVFDIED 155
KG+GK HAKWSP AT Y EP+ + + + EE +L E P VF I+
Sbjct 166 KALARKGIGKDHAKWSPAATVTYMYEPDIIINEEMMNTLTDEEKIDLIESSPTKVFGIDP 225
Query 156 STGRAYAKFPRNCTTCRACLERFENQ-----LQLNKIPDQFIFSIESTGSV 201
TG+ P T +++ E ++++ D F+F++ESTG++
Sbjct 226 VTGQVVVVDPEAYTYDEEVIKKAEAMGKPGLIEIHPKHDSFVFTVESTGAL 276
> Hs14702171
Length=275
Score = 76.6 bits (187), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 48/132 (36%), Positives = 71/132 (53%), Gaps = 9/132 (6%)
Query 1 EVLAHRLGLIPFKVNP--KKLKFRN--PNQELSEENSLKFKMHVKCSPGDLKPHQKSMPV 56
E +AHRLGLIP + KL++ +E E S++F + V+C+ D H S +
Sbjct 62 EFIAHRLGLIPLISDDIVDKLQYSRDCTCEEFCPECSVEFTLDVRCNE-DQTRHVTSRDL 120
Query 57 YARDLKWIPMSERQKQKFAQDPPAPV-HPDILITKLRPGQEIELFGFLEKGLGKTHAKWS 115
+ + IP++ R + DP V DILI KLR GQE+ L + +KG GK HAKW+
Sbjct 121 ISNSPRVIPVTSRNRD---NDPNDYVEQDDILIVKLRKGQELRLRAYAKKGFGKEHAKWN 177
Query 116 PVATAVYRLEPE 127
P A + +P+
Sbjct 178 PTAGVAFEYDPD 189
> 7298103
Length=275
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/131 (32%), Positives = 72/131 (54%), Gaps = 6/131 (4%)
Query 1 EVLAHRLGLIPFKVNP--KKLKFRNPN--QELSEENSLKFKMHVKCSPGDLKPHQKSMPV 56
E LAHR+GLIP + ++L++ + E S++F + VKCS + H + +
Sbjct 62 EFLAHRIGLIPLISDDVVERLQYTRDCICLDFCPECSVEFTLDVKCSEEQTR-HVTTADL 120
Query 57 YARDLKWIPMSERQKQKFAQDPPAPVHPDILITKLRPGQEIELFGFLEKGLGKTHAKWSP 116
+ + K +P++ R Q + + +ILI KLR GQE++L + +KG GK HAKW+P
Sbjct 121 KSSNAKVLPVTSRN-QGEEDNEYGESNDEILIIKLRKGQELKLRAYAKKGFGKEHAKWNP 179
Query 117 VATAVYRLEPE 127
A + +P+
Sbjct 180 TAGVCFEYDPD 190
> CE05370_1
Length=303
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 54/208 (25%), Positives = 94/208 (45%), Gaps = 29/208 (13%)
Query 1 EVLAHRLGLIPF--KVNPKKLKFRNPNQ--ELSEENSLKFKMHVKCSPGDL--KPHQKSM 54
E +AHR+GLIPF + +K+++ + E +E S+ F + +KC + M
Sbjct 62 EFIAHRMGLIPFISDYHVEKMQYTRDCECAEFCDECSIPFILQMKCKDEATLAVTTEHLM 121
Query 55 PVYARDLKWIPMSERQ-KQKFAQDPPAPVHPDILITKLRPGQEIELFGFLEKGLGKTHAK 113
P++ D P + +++ + +ILI KLR GQE+ L +++KG GK HAK
Sbjct 122 PMHNLDTTVRPACGKALRERGSTRDEFHNREEILIVKLRKGQELNLKAYVKKGFGKEHAK 181
Query 114 WSPVATAVYRLEPEFVFTSPIEGEEAKELKELCPMGVFDIEDSTGRAYAKFPRNCTTCRA 173
W+P + +P+ I ++E+ ++ P + T A
Sbjct 182 WNPTCGVAFEYDPDNALRHTIYP---------------NVEEWPRSDHSSLPEDSTEKEA 226
Query 174 CLERFENQLQLNKIPDQFIFSIESTGSV 201
E + P++F FSIE TG++
Sbjct 227 PFEP-------DNKPNKFWFSIEGTGAL 247
> SPCC1442.10c
Length=297
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 43/134 (32%), Positives = 66/134 (49%), Gaps = 18/134 (13%)
Query 1 EVLAHRLGLIPFKVN------PKKLKF-RNPN-QELSEENSLKFKMHVKCSPGDLKPHQK 52
E LAHRLG+IP + P L++ RN + + + S++ ++ KC+ +
Sbjct 60 EFLAHRLGMIPLDSSNIDEPPPVGLEYTRNCDCDQYCPKCSVELFLNAKCT------GEG 113
Query 53 SMPVYARDLKWIPMSERQKQKFAQDPPAPVHPDILITKLRPGQEIELFGFLEKGLGKTHA 112
+M +YARDL + + DP + LI KLR QEI L +KG+ K HA
Sbjct 114 TMEIYARDLV-VSSNSSLGHPILADPKSR---GPLICKLRKEQEISLRCIAKKGIAKEHA 169
Query 113 KWSPVATAVYRLEP 126
KWSP + + +P
Sbjct 170 KWSPTSAVAFEYDP 183
> YIL021w
Length=318
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 38/129 (29%), Positives = 62/129 (48%), Gaps = 10/129 (7%)
Query 1 EVLAHRLGLIPFK-VNPKKLKFRNPNQELSEENSLKFKMHVKCSPGDLKPHQKSMPVYAR 59
E +AHRLGLIP + ++ ++L++ E++ K + + + + VY++
Sbjct 61 EFIAHRLGLIPLQSMDIEQLEYSRDC--FCEDHCDKCSVVLTLQA--FGESESTTNVYSK 116
Query 60 DLKWIP--MSERQKQKFAQDPPAPVHPDILITKLRPGQEIELFGFLEKGLGKTHAKWSPV 117
DL + M QD +LI KLR GQE++L +KG+ K HAKW P
Sbjct 117 DLVIVSNLMGRNIGHPIIQDKEGN---GVLICKLRKGQELKLTCVAKKGIAKEHAKWGPA 173
Query 118 ATAVYRLEP 126
A + +P
Sbjct 174 AAIEFEYDP 182
> ECU06g0600
Length=240
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 44/146 (30%), Positives = 64/146 (43%), Gaps = 28/146 (19%)
Query 1 EVLAHRLGLIPFKVNPKKLKFRN--PNQELSEENSLKFKMHVKCSPGDLKPHQKSMPVYA 58
EVLAHRLGLIP + ++LK++ E S+ F++ V + L+ V
Sbjct 57 EVLAHRLGLIPI-YSKRELKYKEECSCSGFCSECSVVFEIDVVHTDDTLRT------VST 109
Query 59 RDLKWIPMSERQKQKFAQDPPAPVHPDILITKLRPGQEIELFGFLEKGLGKTHAKWSPVA 118
RD+ + + +ITKL Q ++ KG+GKTHAKWSPV+
Sbjct 110 RDI------------VCDEEDVIIKSGPVITKLARDQGLKARCIGRKGVGKTHAKWSPVS 157
Query 119 TAVY-------RLEPEFVFTSPIEGE 137
+ R E + F IE E
Sbjct 158 AISFEYDKDNVRRETNYWFEESIEEE 183
> At3g14810
Length=853
Score = 34.3 bits (77), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 20/44 (45%), Positives = 25/44 (56%), Gaps = 3/44 (6%)
Query 65 PMSERQKQKF-AQDPPAPVHPDILITKLRPGQEIELF--GFLEK 105
PM R K K QDPP P HP I T+++ G+ +F GFL K
Sbjct 187 PMLSRNKTKSRLQDPPTPTHPAIDKTEMKSGRRSGIFKSGFLGK 230
> Hs14916465
Length=635
Score = 30.4 bits (67), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 19/48 (39%), Positives = 25/48 (52%), Gaps = 2/48 (4%)
Query 4 AHRLGLIPFKVNPKKLKFRNPNQELSEENSLKFKMHVKCSPGDLKPHQ 51
HRL K N K+ K +N + EENSL+F + +PGD P Q
Sbjct 28 GHRLSKTKQKRNRKRNKKQNSQNRIMEENSLEFLSDL--TPGDQDPSQ 73
> At3g49080
Length=143
Score = 30.4 bits (67), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 155 DSTGRAYAKFPRNCTTCRACLERFENQLQLN 185
D TGRAY R C+ R ++ E + Q+N
Sbjct 14 DETGRAYGTGRRKCSIARVWIQPGEGKFQVN 44
> Hs5031825
Length=685
Score = 29.6 bits (65), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 20/52 (38%), Gaps = 16/52 (30%)
Query 158 GRAYAKFPRN----------------CTTCRACLERFENQLQLNKIPDQFIF 193
GR+Y + N CT C CLE FEN L +P +F
Sbjct 592 GRSYGSYNTNEDMEPDWLTWPADMLHCTECVVCLENFENGCLLMGLPCGHVF 643
Lambda K H
0.319 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3515233148
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40